Filter
Associated Lab
- Aso Lab (8) Apply Aso Lab filter
- Branson Lab (1) Apply Branson Lab filter
- Card Lab (4) Apply Card Lab filter
- Cardona Lab (2) Apply Cardona Lab filter
- Darshan Lab (1) Apply Darshan Lab filter
- Dickson Lab (4) Apply Dickson Lab filter
- Espinosa Medina Lab (2) Apply Espinosa Medina Lab filter
- Fitzgerald Lab (1) Apply Fitzgerald Lab filter
- Funke Lab (1) Apply Funke Lab filter
- Heberlein Lab (1) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Jayaraman Lab (2) Apply Jayaraman Lab filter
- Looger Lab (1) Apply Looger Lab filter
- Reiser Lab (6) Apply Reiser Lab filter
- Romani Lab (2) Apply Romani Lab filter
- Rubin Lab (8) Apply Rubin Lab filter
- Stern Lab (3) Apply Stern Lab filter
- Truman Lab (2) Apply Truman Lab filter
- Turner Lab (4) Apply Turner Lab filter
- Zlatic Lab (1) Apply Zlatic Lab filter
Associated Project Team
Associated Support Team
- Remove Fly Facility filter Fly Facility
- Janelia Experimental Technology (6) Apply Janelia Experimental Technology filter
- Molecular Genomics (3) Apply Molecular Genomics filter
- Project Technical Resources (13) Apply Project Technical Resources filter
- Quantitative Genomics (4) Apply Quantitative Genomics filter
- Scientific Computing Software (3) Apply Scientific Computing Software filter
39 Janelia Publications
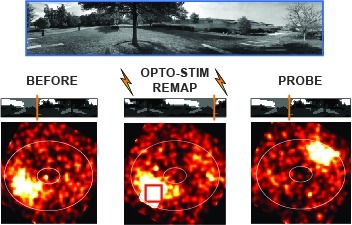
Showing 31-39 of 39 resultsMany animals rely on an internal heading representation when navigating in varied environments. How this representation is linked to the sensory cues that define different surroundings is unclear. In the fly brain, heading is represented by 'compass' neurons that innervate a ring-shaped structure known as the ellipsoid body. Each compass neuron receives inputs from 'ring' neurons that are selective for particular visual features; this combination provides an ideal substrate for the extraction of directional information from a visual scene. Here we combine two-photon calcium imaging and optogenetics in tethered flying flies with circuit modelling, and show how the correlated activity of compass and visual neurons drives plasticity, which flexibly transforms two-dimensional visual cues into a stable heading representation. We also describe how this plasticity enables the fly to convert a partial heading representation, established from orienting within part of a novel setting, into a complete heading representation. Our results provide mechanistic insight into the memory-related computations that are essential for flexible navigation in varied surroundings.
Animals employ diverse learning rules and synaptic plasticity dynamics to record temporal and statistical information about the world. However, the molecular mechanisms underlying this diversity are poorly understood. The anatomically defined compartments of the insect mushroom body function as parallel units of associative learning, with different learning rates, memory decay dynamics and flexibility (Aso & Rubin 2016). Here we show that nitric oxide (NO) acts as a neurotransmitter in a subset of dopaminergic neurons in . NO's effects develop more slowly than those of dopamine and depend on soluble guanylate cyclase in postsynaptic Kenyon cells. NO acts antagonistically to dopamine; it shortens memory retention and facilitates the rapid updating of memories. The interplay of NO and dopamine enables memories stored in local domains along Kenyon cell axons to be specialized for predicting the value of odors based only on recent events. Our results provide key mechanistic insights into how diverse memory dynamics are established in parallel memory systems.
Animals perform or terminate particular behaviors by integrating external cues and internal states through neural circuits. Identifying neural substrates and their molecular modulators promoting or inhibiting animal behaviors are key steps to understand how neural circuits control behaviors. Here, we identify the Cholecystokinin-like peptide Drosulfakinin (DSK) that functions at single-neuron resolution to suppress male sexual behavior in Drosophila. We found that Dsk neurons physiologically interact with male-specific P1 neurons, part of a command center for male sexual behaviors, and function oppositely to regulate multiple arousal-related behaviors including sex, sleep and spontaneous walking. We further found that the DSK-2 peptide functions through its receptor CCKLR-17D3 to suppress sexual behaviors in flies. Such a neuropeptide circuit largely overlaps with the fruitless-expressing neural circuit that governs most aspects of male sexual behaviors. Thus DSK/CCKLR signaling in the sex circuitry functions antagonistically with P1 neurons to balance arousal levels and modulate sexual behaviors.
males perform a series of courtship behaviors that, when successful, result in copulation with a female. For over a century, mutations in the gene, named for its effects on pigmentation, have been known to reduce male mating success. Prior work has suggested that influences mating behavior through effects on wing extension, song, and/or courtship vigor. Here, we rule out these explanations, as well as effects on the nervous system more generally, and find instead that the effects of on male mating success are mediated by its effects on pigmentation of male-specific leg structures called sex combs. Loss of expression in these modified bristles reduces their melanization, which changes their structure and causes difficulty grasping females prior to copulation. These data illustrate why the mechanical properties of anatomy, not just neural circuitry, must be considered to fully understand the development and evolution of behavior.
We present CLADES (Cell Lineage Access Driven by an Edition Sequence), a technology for cell lineage studies based on CRISPR/Cas9. CLADES relies on a system of genetic switches to activate and inactivate reporter genes in a pre-determined order. Targeting CLADES to progenitor cells allows the progeny to inherit a sequential cascade of reporters, coupling birth order with reporter expression. This gives us temporal resolution of lineage development that can be used to deconstruct an extended cell lineage by tracking the reporters expressed in the progeny. When targeted to the germ line, the same cascade progresses across animal generations, marking each generation with the corresponding combination of reporters. CLADES thus offers an innovative strategy for making programmable cascades of genes that can be used for genetic manipulation or to record serial biological events.
Optogenetics is possibly the most revolutionary advance in neuroscience research techniques within the last decade. Here, we describe lab modules, presented at a workshop for undergraduate neuroscience educators, using optogenetic control of neurons in the fruit fly Drosophila melanogaster. Drosophila is a genetically accessible model system that combines behavioral and neurophysiological complexity, ease of use, and high research relevance. One lab module utilized two transgenic Drosophila strains, each activating specific circuits underlying startle behavior and backwards locomotion, respectively. The red-shifted channelrhodopsin, CsChrimson, was expressed in neurons sharing a common transcriptional profile, with the expression pattern further refined by the use of a Split GAL4 intersectional activation system. Another set of strains was used to investigate synaptic transmission at the larval neuromuscular junction. These expressed Channelrhodopsin 2 (ChR2) in glutamatergic neurons, including the motor neurons. The first strain expressed ChR2 in a wild type background, while the second contained the SNAP-25ts mutant allele, which confers heightened evoked potential amplitude and greatly increased spontaneous vesicle release frequency at the larval neuromuscular junction. These modules introduced educators and students to the use of optogenetic stimulation to control behavior and evoked release at a model synapse, and establish a basis for students to explore neurophysiology using this technique, through recapitulating classic experiments and conducting independent research.
The ability to reproducibly target expression of transgenes to small, defined subsets of cells is a key experimental tool for understanding many biological processes. The Drosophila nervous system contains thousands of distinct cell types and it has generally not been possible to limit expression to one or a few cell types when using a single segment of genomic DNA as an enhancer to drive expression. Intersectional methods, in which expression of the transgene only occurs where two different enhancers overlap in their expression patterns, can be used to achieve the desired specificity. This report describes a set of over 2,800 transgenic lines for use with the split-GAL4 intersectional method.
To facilitate large scale functional studies in Drosophila, the Drosophila Transgenic RNAi Project (TRiP) at Harvard Medical School (HMS) was established along with several goals: developing efficient vectors for RNAi that work in all tissues, generating a genome scale collection of RNAi stocks with input from the community, distributing the lines as they are generated through existing stock centers, validating as many lines as possible using RT-qPCR and phenotypic analyses, and developing tools and web resources for identifying RNAi lines and retrieving existing information on their quality. With these goals in mind, here we describe in detail the various tools we developed and the status of the collection, which is currently comprised of 11,491 lines and covering 71% of Drosophila genes. Data on the characterization of the lines either by RT-qPCR or phenotype is available on a dedicated web site, the RNAi Stock Validation and Phenotypes Project (RSVP; www.flyrnai.org/RSVP.html), and stocks are available from three stock centers, the Bloomington Drosophila Stock Center (USA), National Institute of Genetics (Japan), and TsingHua Fly Center (China).
Conditional expression of hairpin constructs in Drosophila is a powerful method to disrupt the activity of single genes with a spatial and temporal resolution that is impossible, or exceedingly difficult, using classical genetic methods. We previously described a method (Ni et al. 2008) whereby RNAi constructs are targeted into the genome by the phiC31-mediated integration approach using Vermilion-AttB-Loxp-Intron-UAS-MCS (VALIUM), a vector that contains vermilion as a selectable marker, an attB sequence to allow for phiC31-targeted integration at genomic attP landing sites, two pentamers of UAS, the hsp70 core promoter, a multiple cloning site, and two introns. As the level of gene activity knockdown associated with transgenic RNAi depends on the level of expression of the hairpin constructs, we generated a number of derivatives of our initial vector, called the "VALIUM" series, to improve the efficiency of the method. Here, we report the results from the systematic analysis of these derivatives and characterize VALIUM10 as the most optimal vector of this series. A critical feature of VALIUM10 is the presence of gypsy insulator sequences that boost dramatically the level of knockdown. We document the efficacy of VALIUM as a vector to analyze the phenotype of genes expressed in the nervous system and have generated a library of 2282 constructs targeting 2043 genes that will be particularly useful for studies of the nervous system as they target, in particular, transcription factors, ion channels, and transporters.