Filter
Associated Lab
- Betzig Lab (8) Apply Betzig Lab filter
- Eddy/Rivas Lab (2) Apply Eddy/Rivas Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Hess Lab (2) Apply Hess Lab filter
- Ji Lab (4) Apply Ji Lab filter
- Looger Lab (3) Apply Looger Lab filter
- Magee Lab (4) Apply Magee Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Rubin Lab (2) Apply Rubin Lab filter
- Schreiter Lab (1) Apply Schreiter Lab filter
- Shroff Lab (6) Apply Shroff Lab filter
- Spruston Lab (3) Apply Spruston Lab filter
- Sternson Lab (1) Apply Sternson Lab filter
- Svoboda Lab (6) Apply Svoboda Lab filter
- Truman Lab (1) Apply Truman Lab filter
Publication Date
- December 2008 (8) Apply December 2008 filter
- November 2008 (1) Apply November 2008 filter
- October 2008 (1) Apply October 2008 filter
- September 2008 (2) Apply September 2008 filter
- August 2008 (3) Apply August 2008 filter
- July 2008 (4) Apply July 2008 filter
- June 2008 (1) Apply June 2008 filter
- May 2008 (3) Apply May 2008 filter
- April 2008 (1) Apply April 2008 filter
- March 2008 (5) Apply March 2008 filter
- February 2008 (5) Apply February 2008 filter
- January 2008 (6) Apply January 2008 filter
- Remove 2008 filter 2008
40 Janelia Publications
Showing 21-30 of 40 resultsSequence database searches require accurate estimation of the statistical significance of scores. Optimal local sequence alignment scores follow Gumbel distributions, but determining an important parameter of the distribution (lambda) requires time-consuming computational simulation. Moreover, optimal alignment scores are less powerful than probabilistic scores that integrate over alignment uncertainty ("Forward" scores), but the expected distribution of Forward scores remains unknown. Here, I conjecture that both expected score distributions have simple, predictable forms when full probabilistic modeling methods are used. For a probabilistic model of local sequence alignment, optimal alignment bit scores ("Viterbi" scores) are Gumbel-distributed with constant lambda = log 2, and the high scoring tail of Forward scores is exponential with the same constant lambda. Simulation studies support these conjectures over a wide range of profile/sequence comparisons, using 9,318 profile-hidden Markov models from the Pfam database. This enables efficient and accurate determination of expectation values (E-values) for both Viterbi and Forward scores for probabilistic local alignments.
Over hundreds of millions of years, evolution has optimized brain design to maximize its functionality while minimizing costs associated with building and maintenance. This observation suggests that one can use optimization theory to rationalize various features of brain design. Here, we attempt to explain the dimensions and branching structure of dendritic arbors by minimizing dendritic cost for given potential synaptic connectivity. Assuming only that dendritic cost increases with total dendritic length and path length from synapses to soma, we find that branching, planar, and compact dendritic arbors, such as those belonging to Purkinje cells in the cerebellum, are optimal. The theory predicts that adjacent Purkinje dendritic arbors should spatially segregate. In addition, we propose two explicit cost function expressions, falsifiable by measuring dendritic caliber near bifurcations.
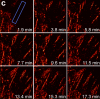
We demonstrate live-cell super-resolution imaging using photoactivated localization microscopy (PALM). The use of photon-tolerant cell lines in combination with the high resolution and molecular sensitivity of PALM permitted us to investigate the nanoscale dynamics within individual adhesion complexes (ACs) in living cells under physiological conditions for as long as 25 min, with half of the time spent collecting the PALM images at spatial resolutions down to approximately 60 nm and frame rates as short as 25 s. We visualized the formation of ACs and measured the fractional gain and loss of individual paxillin molecules as each AC evolved. By allowing observation of a wide variety of nanoscale dynamics, live-cell PALM provides insights into molecular assembly during the initiation, maturation and dissolution of cellular processes.
Commentary: The first example of true live cell and time lapse imaging by localization microscopy (as opposed to particle tracking), this paper uses the Nyquist criterion to establish a necessary condition for true spatial resolution based on the density of localized molecules – a condition often unmet in claims elsewhere in the superresolution literature.
By any method, higher spatiotemporal resolution requires increasing light exposure at the specimen, making noninvasive imaging increasingly difficult. Here, simultaneous differential interference contrast imaging is used to establish that cells behave physiologically before, during, and after PALM imaging. Similar controls are lacking from many supposed “live cell” superresolution demonstrations.
The development of high-resolution microscopy makes possible the high-throughput screening of cellular information, such as gene expression at single cell resolution. One of the critical enabling techniques yet to be developed is the automatic recognition or annotation of specific cells in a 3D image stack. In this paper, we present a novel graph-based algorithm, ARC, that determines cell identities in a 3D confocal image of C. elegans based on their highly stereotyped arrangement. This is an essential step in our work on gene expression analysis of C. elegans at the resolution of single cells. Our ARC method integrates both the absolute and relative spatial locations of cells in a C. elegans body. It uses a marker-guided, spatially-constrained, two-stage bipartite matching to find the optimal match between cells in a subject image and cells in 15 template images that have been manually annotated and vetted. We applied ARC to the recognition of cells in 3D confocal images of the first larval stage (L1) of C. elegans hermaphrodites, and achieved an average accuracy of 94.91%.
Although information storage in the central nervous system is thought to be primarily mediated by various forms of synaptic plasticity, other mechanisms, such as modifications in membrane excitability, are available. Local dendritic spikes are nonlinear voltage events that are initiated within dendritic branches by spatially clustered and temporally synchronous synaptic input. That local spikes selectively respond only to appropriately correlated input allows them to function as input feature detectors and potentially as powerful information storage mechanisms. However, it is currently unknown whether any effective form of local dendritic spike plasticity exists. Here we show that the coupling between local dendritic spikes and the soma of rat hippocampal CA1 pyramidal neurons can be modified in a branch-specific manner through an N-methyl-d-aspartate receptor (NMDAR)-dependent regulation of dendritic Kv4.2 potassium channels. These data suggest that compartmentalized changes in branch excitability could store multiple complex features of synaptic input, such as their spatio-temporal correlation. We propose that this ’branch strength potentiation’ represents a previously unknown form of information storage that is distinct from that produced by changes in synaptic efficacy both at the mechanistic level and in the type of information stored.
Genetically-encoded calcium indicators (GECIs) hold the promise of monitoring [Ca(2+)] in selected populations of neurons and in specific cellular compartments. Relating GECI fluorescence to neuronal activity requires quantitative characterization. We have characterized a promising new genetically-encoded calcium indicator-GCaMP2-in mammalian pyramidal neurons. Fluorescence changes in response to single action potentials (17+/-10% DeltaF/F [mean+/-SD]) could be detected in some, but not all, neurons. Trains of high-frequency action potentials yielded robust responses (302+/-50% for trains of 40 action potentials at 83 Hz). Responses were similar in acute brain slices from in utero electroporated mice, indicating that long-term expression did not interfere with GCaMP2 function. Membrane-targeted versions of GCaMP2 did not yield larger signals than their non-targeted counterparts. We further targeted GCaMP2 to dendritic spines to monitor Ca(2+) accumulations evoked by activation of synaptic NMDA receptors. We observed robust DeltaF/F responses (range: 37%-264%) to single spine uncaging stimuli that were correlated with NMDA receptor currents measured through a somatic patch pipette. One major drawback of GCaMP2 was its low baseline fluorescence. Our results show that GCaMP2 is improved from the previous versions of GCaMP and may be suited to detect bursts of high-frequency action potentials and synaptic currents in vivo.
Understanding the principles of information processing in neural circuits requires systematic characterization of the participating cell types and their connections, and the ability to measure and perturb their activity. Genetic approaches promise to bring experimental access to complex neural systems, including genetic stalwarts such as the fly and mouse, but also to nongenetic systems such as primates. Together with anatomical and physiological methods, cell-type-specific expression of protein markers and sensors and transducers will be critical to construct circuit diagrams and to measure the activity of genetically defined neurons. Inactivation and activation of genetically defined cell types will establish causal relationships between activity in specific groups of neurons, circuit function, and animal behavior. Genetic analysis thus promises to reveal the logic of the neural circuits in complex brains that guide behaviors. Here we review progress in the genetic analysis of neural circuits and discuss directions for future research and development.
Pyramidal neurons are characterized by their distinct apical and basal dendritic trees and the pyramidal shape of their soma. They are found in several regions of the CNS and, although the reasons for their abundance remain unclear, functional studies--especially of CA1 hippocampal and layer V neocortical pyramidal neurons--have offered insights into the functions of their unique cellular architecture. Pyramidal neurons are not all identical, but some shared functional principles can be identified. In particular, the existence of dendritic domains with distinct synaptic inputs, excitability, modulation and plasticity appears to be a common feature that allows synapses throughout the dendritic tree to contribute to action-potential generation. These properties support a variety of coincidence-detection mechanisms, which are likely to be crucial for synaptic integration and plasticity.
The identification of synaptic partners is challenging in dense nerve bundles, where many processes occupy regions beneath the resolution of conventional light microscopy. To address this difficulty, we have developed GRASP, a system to label membrane contacts and synapses between two cells in living animals. Two complementary fragments of GFP are expressed on different cells, tethered to extracellular domains of transmembrane carrier proteins. When the complementary GFP fragments are fused to ubiquitous transmembrane proteins, GFP fluorescence appears uniformly along membrane contacts between the two cells. When one or both GFP fragments are fused to synaptic transmembrane proteins, GFP fluorescence is tightly localized to synapses. GRASP marks known synaptic contacts in C. elegans, correctly identifies changes in mutants with altered synaptic specificity, and can uncover new information about synaptic locations as confirmed by electron microscopy. GRASP may prove particularly useful for defining connectivity in complex nervous systems.