Filter
Associated Lab
Publication Date
3 Janelia Publications
Showing 1-3 of 3 resultsSequence database searches require accurate estimation of the statistical significance of scores. Optimal local sequence alignment scores follow Gumbel distributions, but determining an important parameter of the distribution (lambda) requires time-consuming computational simulation. Moreover, optimal alignment scores are less powerful than probabilistic scores that integrate over alignment uncertainty ("Forward" scores), but the expected distribution of Forward scores remains unknown. Here, I conjecture that both expected score distributions have simple, predictable forms when full probabilistic modeling methods are used. For a probabilistic model of local sequence alignment, optimal alignment bit scores ("Viterbi" scores) are Gumbel-distributed with constant lambda = log 2, and the high scoring tail of Forward scores is exponential with the same constant lambda. Simulation studies support these conjectures over a wide range of profile/sequence comparisons, using 9,318 profile-hidden Markov models from the Pfam database. This enables efficient and accurate determination of expectation values (E-values) for both Viterbi and Forward scores for probabilistic local alignments.
Over hundreds of millions of years, evolution has optimized brain design to maximize its functionality while minimizing costs associated with building and maintenance. This observation suggests that one can use optimization theory to rationalize various features of brain design. Here, we attempt to explain the dimensions and branching structure of dendritic arbors by minimizing dendritic cost for given potential synaptic connectivity. Assuming only that dendritic cost increases with total dendritic length and path length from synapses to soma, we find that branching, planar, and compact dendritic arbors, such as those belonging to Purkinje cells in the cerebellum, are optimal. The theory predicts that adjacent Purkinje dendritic arbors should spatially segregate. In addition, we propose two explicit cost function expressions, falsifiable by measuring dendritic caliber near bifurcations.
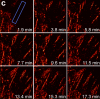
We demonstrate live-cell super-resolution imaging using photoactivated localization microscopy (PALM). The use of photon-tolerant cell lines in combination with the high resolution and molecular sensitivity of PALM permitted us to investigate the nanoscale dynamics within individual adhesion complexes (ACs) in living cells under physiological conditions for as long as 25 min, with half of the time spent collecting the PALM images at spatial resolutions down to approximately 60 nm and frame rates as short as 25 s. We visualized the formation of ACs and measured the fractional gain and loss of individual paxillin molecules as each AC evolved. By allowing observation of a wide variety of nanoscale dynamics, live-cell PALM provides insights into molecular assembly during the initiation, maturation and dissolution of cellular processes.
Commentary: The first example of true live cell and time lapse imaging by localization microscopy (as opposed to particle tracking), this paper uses the Nyquist criterion to establish a necessary condition for true spatial resolution based on the density of localized molecules – a condition often unmet in claims elsewhere in the superresolution literature.
By any method, higher spatiotemporal resolution requires increasing light exposure at the specimen, making noninvasive imaging increasingly difficult. Here, simultaneous differential interference contrast imaging is used to establish that cells behave physiologically before, during, and after PALM imaging. Similar controls are lacking from many supposed “live cell” superresolution demonstrations.