Filter
Associated Lab
- Ahrens Lab (1) Apply Ahrens Lab filter
- Aso Lab (2) Apply Aso Lab filter
- Baker Lab (2) Apply Baker Lab filter
- Betzig Lab (5) Apply Betzig Lab filter
- Bock Lab (1) Apply Bock Lab filter
- Branson Lab (3) Apply Branson Lab filter
- Card Lab (1) Apply Card Lab filter
- Cardona Lab (6) Apply Cardona Lab filter
- Chklovskii Lab (1) Apply Chklovskii Lab filter
- Cui Lab (6) Apply Cui Lab filter
- Dickson Lab (3) Apply Dickson Lab filter
- Druckmann Lab (3) Apply Druckmann Lab filter
- Eddy/Rivas Lab (1) Apply Eddy/Rivas Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Fitzgerald Lab (1) Apply Fitzgerald Lab filter
- Gonen Lab (7) Apply Gonen Lab filter
- Grigorieff Lab (7) Apply Grigorieff Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (9) Apply Heberlein Lab filter
- Hess Lab (3) Apply Hess Lab filter
- Jayaraman Lab (2) Apply Jayaraman Lab filter
- Ji Lab (2) Apply Ji Lab filter
- Kainmueller Lab (2) Apply Kainmueller Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keleman Lab (2) Apply Keleman Lab filter
- Keller Lab (3) Apply Keller Lab filter
- Koay Lab (1) Apply Koay Lab filter
- Lavis Lab (4) Apply Lavis Lab filter
- Lee (Albert) Lab (2) Apply Lee (Albert) Lab filter
- Leonardo Lab (2) Apply Leonardo Lab filter
- Lippincott-Schwartz Lab (12) Apply Lippincott-Schwartz Lab filter
- Looger Lab (13) Apply Looger Lab filter
- Magee Lab (6) Apply Magee Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Pachitariu Lab (1) Apply Pachitariu Lab filter
- Pastalkova Lab (1) Apply Pastalkova Lab filter
- Pavlopoulos Lab (1) Apply Pavlopoulos Lab filter
- Pedram Lab (1) Apply Pedram Lab filter
- Reiser Lab (1) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Rubin Lab (8) Apply Rubin Lab filter
- Saalfeld Lab (7) Apply Saalfeld Lab filter
- Satou Lab (2) Apply Satou Lab filter
- Scheffer Lab (3) Apply Scheffer Lab filter
- Schreiter Lab (2) Apply Schreiter Lab filter
- Sgro Lab (1) Apply Sgro Lab filter
- Simpson Lab (1) Apply Simpson Lab filter
- Singer Lab (11) Apply Singer Lab filter
- Spruston Lab (4) Apply Spruston Lab filter
- Stern Lab (5) Apply Stern Lab filter
- Sternson Lab (4) Apply Sternson Lab filter
- Svoboda Lab (9) Apply Svoboda Lab filter
- Tervo Lab (1) Apply Tervo Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (3) Apply Truman Lab filter
Associated Project Team
Publication Date
- December 2012 (16) Apply December 2012 filter
- November 2012 (16) Apply November 2012 filter
- October 2012 (23) Apply October 2012 filter
- September 2012 (6) Apply September 2012 filter
- August 2012 (13) Apply August 2012 filter
- July 2012 (9) Apply July 2012 filter
- June 2012 (15) Apply June 2012 filter
- May 2012 (13) Apply May 2012 filter
- April 2012 (14) Apply April 2012 filter
- March 2012 (10) Apply March 2012 filter
- February 2012 (19) Apply February 2012 filter
- January 2012 (36) Apply January 2012 filter
- Remove 2012 filter 2012
Type of Publication
190 Publications
Showing 181-190 of 190 resultsPhylogenetic footprinting has revealed that cis-regulatory enhancers consist of conserved DNA sequence clusters (CSCs). Currently, there is no systematic approach for enhancer discovery and analysis that takes full-advantage of the sequence information within enhancer CSCs.
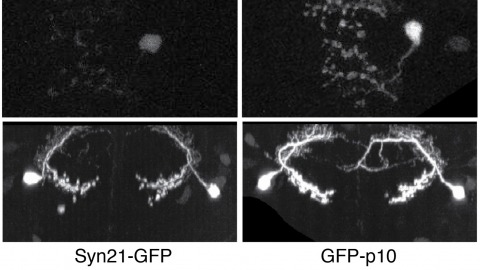
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibire(ts1), and for the use of cytoplasmic GFP to image the fine processes of neurons.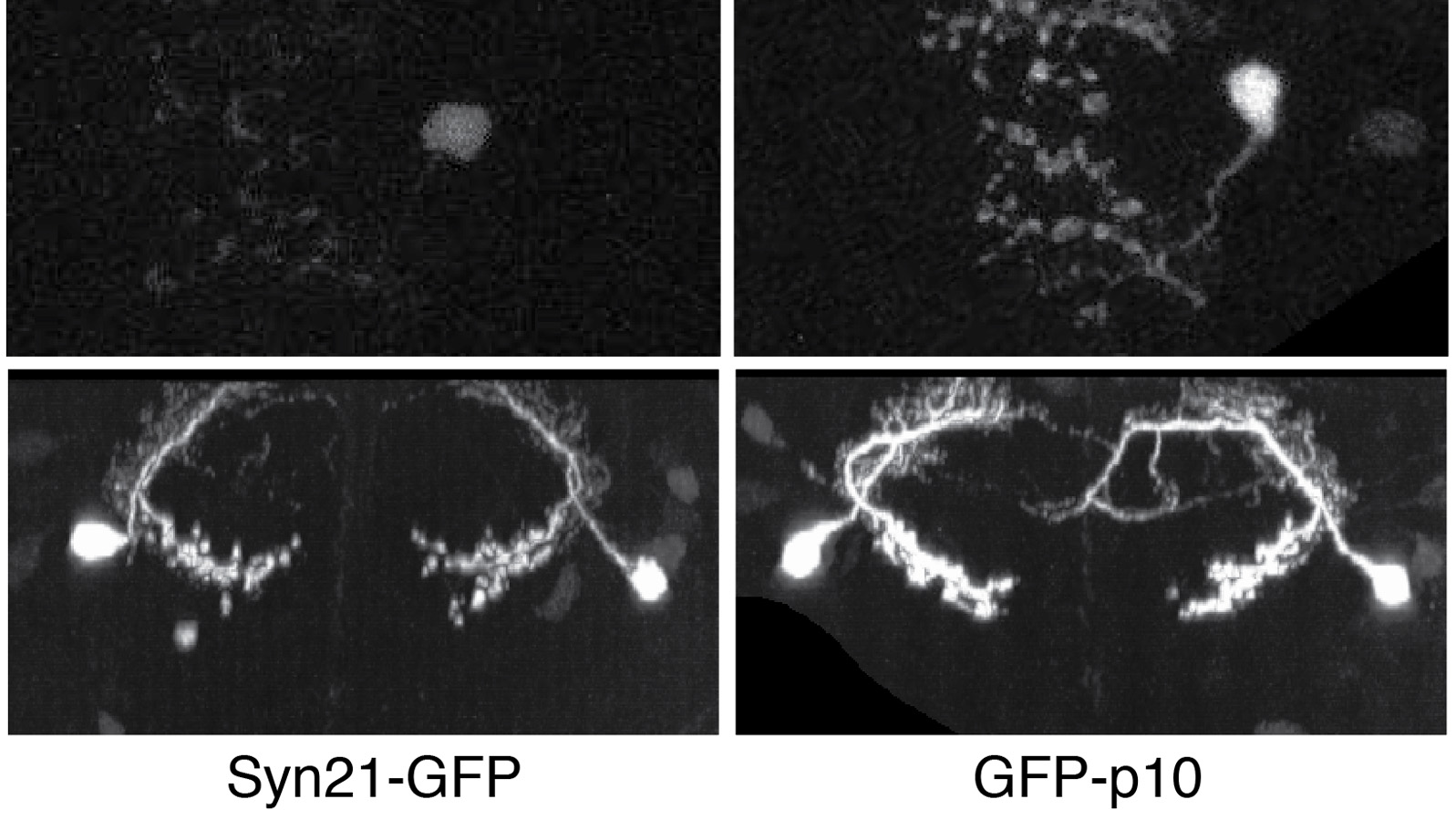
The visual neurons of many animals process sensory input differently depending on the animal’s state of locomotion. Now, new work in Drosophila melanogaster shows that neuromodulatory neurons active during flight boost responses of neurons in the visual system.
In a wide range of biological studies, it is highly desirable to visualize and analyze three-dimensional (3D) microscopic images. In this primer, we first introduce several major methods for visualizing typical 3D images and related multi-scale, multi-time-point, multi-color data sets. Then, we discuss three key categories of image analysis tasks, namely segmentation, registration, and annotation. We demonstrate how to pipeline these visualization and analysis modules using examples of profiling the single-cell gene-expression of C. elegans and constructing a map of stereotyped neurite tracts in a fruit fly brain.
Methods useful for exploring the formation and functions of primary cilia in living cells are described here. First, multiple protocols for visualizing solitary cilia that extend away from the cell body are described. Primary cilia collect, synthesize, and transmit information about the extracellular space into the cell body to promote critical cellular responses. Problems with cilia formation or function can lead to dramatic changes in cell physiology. These methods can be used to assess cilia formation and length, the location of the cilium relative to other cellular structures, and localization of specific proteins to the cilium. The subsequent protocols describe how to quantify movement of fluorescent molecules within the cilium using kymographs, photobleaching, and photoconversion. The microtubules that form the structural scaffold of the cilium are also critical avenues for kinesin and dynein-mediated movement of proteins within the cilium. Assessing intraflagellar dynamics can provide insight into mechanisms of ciliary-mediated signal perception and transmission.
Methods useful for exploring the formation and functions of primary cilia in living cells are described here. First, multiple protocols for visualizing solitary cilia that extend away from the cell body are described. Primary cilia collect, synthesize, and transmit information about the extracellular space into the cell body to promote critical cellular responses. Problems with cilia formation or function can lead to dramatic changes in cell physiology. These methods can be used to assess cilia formation and length, the location of the cilium relative to other cellular structures, and localization of specific proteins to the cilium. The subsequent protocols describe how to quantify movement of fluorescent molecules within the cilium using kymographs, photobleaching, and photoconversion. The microtubules that form the structural scaffold of the cilium are also critical avenues for kinesin and dynein-mediated movement of proteins within the cilium. Assessing intraflagellar dynamics can provide insight into mechanisms of ciliary-mediated signal perception and transmission.
Fundamental to the success of cell and developmental biology is the ability to tease apart molecular organization in cells and tissues by localizing specific proteins with respect to one another in a native cellular context. However, many key cellular structures (from mitochondrial cristae to nuclear pores) lie below the diffraction limit of visible light, precluding analysis of their organization by conventional approaches. Point-localization superresolution microscopy techniques, such as PALM and STORM, are poised to resolve, with unprecedented clarity, the organizational principles of macromolecular complexes within cells, thus leading to deeper insights into cellular function in both health and disease.
The last decade has seen a rapid increase in the number of tools to acquire volume electron microscopy (EM) data. Several new scanning EM (SEM) imaging methods have emerged, and classical transmission EM (TEM) methods are being scaled up and automated. Here we summarize the new methods for acquiring large EM volumes, and discuss the tradeoffs in terms of resolution, acquisition speed, and reliability. We then assess each method’s applicability to the problem of reconstructing anatomical connectivity between neurons, considering both the current capabilities and future prospects of the method. Finally, we argue that neuronal ’wiring diagrams’ are likely necessary, but not sufficient, to understand the operation of most neuronal circuits: volume EM imaging will likely find its best application in combination with other methods in neuroscience, such as molecular biology, optogenetics, and physiology.
Directed cell motility is at the basis of biological phenomena such as development, wound healing, and metastasis. It has been shown that substrate attachments mediate motility by coupling the cell's cytoskeleton with force generation. However, it has been unclear how the persistence of cell directionality is facilitated. We show that mRNA localization plays an important role in this process, but the mechanism of action is still unknown. In this study, we show that the zipcode-binding protein 1 transports β-actin mRNA to the focal adhesion compartment, where it dwells for minutes, suggesting a means for associating its localization with motility through the formation of stable connections between adhesions and newly synthesized actin filaments. In order to demonstrate this, we developed an approach for assessing the functional consequences of β-actin mRNA and protein localization by tethering the mRNA to a specific location-in this case, the focal adhesion complex. This approach will have a significant impact on cell biology because it is now possible to forcibly direct any mRNA and its cognate protein to specific locations in the cell. This will reveal the importance of localized protein translation on various cellular processes.
β-secretase (or BACE1) is the key enzyme in the production of β-amyloid (Aβ), which accumulates in the senile plaques characteristic for Alzheimer's disease. Consequently, the lack of BACE1 prevents β-processing of the amyloid precursor protein and Aβ production, which made it a promising target for drug development. However, the loss of BACE1 is also detrimental, leading to myelination defects and altered neuronal activity, functions that have been associated with the cleavage of Neuregulin and a voltage-gated sodium channel subunit. Here we show that the Drosophila ortholog of BACE, dBACE, is required for glial survival. Cell-specific knockdown experiments reveal that this is a non-cell autonomous function, as a knockdown of dBACE in photoreceptor neurons leads to progressive degeneration of glia in their target zone, the lamina. Interestingly, this phenotype is suppressed by the loss of the fly amyloid precursor protein (APPL), whereas a secretion-deficient form of APPL enhances the degeneration. This shows that full-length APPL in neurons promotes the death of neighboring glial cells and that β-processing of APPL is needed to prevent glial death. These results therefore not only demonstrate a novel function for an APP protein in glia, but they also show this function specifically requires regulation by β-cleavage.