Filter
Associated Lab
- Aguilera Castrejon Lab (14) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (11) Apply Ahrens Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (12) Apply Betzig Lab filter
- Beyene Lab (5) Apply Beyene Lab filter
- Bock Lab (3) Apply Bock Lab filter
- Branson Lab (3) Apply Branson Lab filter
- Card Lab (6) Apply Card Lab filter
- Cardona Lab (19) Apply Cardona Lab filter
- Chklovskii Lab (3) Apply Chklovskii Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Darshan Lab (4) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (14) Apply Dickson Lab filter
- Druckmann Lab (4) Apply Druckmann Lab filter
- Dudman Lab (12) Apply Dudman Lab filter
- Egnor Lab (7) Apply Egnor Lab filter
- Espinosa Medina Lab (4) Apply Espinosa Medina Lab filter
- Fetter Lab (10) Apply Fetter Lab filter
- Fitzgerald Lab (13) Apply Fitzgerald Lab filter
- Gonen Lab (32) Apply Gonen Lab filter
- Grigorieff Lab (28) Apply Grigorieff Lab filter
- Harris Lab (10) Apply Harris Lab filter
- Heberlein Lab (81) Apply Heberlein Lab filter
- Hermundstad Lab (4) Apply Hermundstad Lab filter
- Hess Lab (3) Apply Hess Lab filter
- Jayaraman Lab (4) Apply Jayaraman Lab filter
- Johnson Lab (5) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (1) Apply Karpova Lab filter
- Keleman Lab (5) Apply Keleman Lab filter
- Keller Lab (15) Apply Keller Lab filter
- Koay Lab (16) Apply Koay Lab filter
- Lavis Lab (12) Apply Lavis Lab filter
- Lee (Albert) Lab (5) Apply Lee (Albert) Lab filter
- Leonardo Lab (4) Apply Leonardo Lab filter
- Li Lab (24) Apply Li Lab filter
- Lippincott-Schwartz Lab (72) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (5) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (5) Apply Liu (Zhe) Lab filter
- Looger Lab (1) Apply Looger Lab filter
- Magee Lab (18) Apply Magee Lab filter
- Menon Lab (6) Apply Menon Lab filter
- Murphy Lab (7) Apply Murphy Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (12) Apply Otopalik Lab filter
- Pachitariu Lab (12) Apply Pachitariu Lab filter
- Pastalkova Lab (13) Apply Pastalkova Lab filter
- Pavlopoulos Lab (12) Apply Pavlopoulos Lab filter
- Pedram Lab (11) Apply Pedram Lab filter
- Reiser Lab (6) Apply Reiser Lab filter
- Riddiford Lab (24) Apply Riddiford Lab filter
- Romani Lab (12) Apply Romani Lab filter
- Rubin Lab (38) Apply Rubin Lab filter
- Saalfeld Lab (17) Apply Saalfeld Lab filter
- Satou Lab (15) Apply Satou Lab filter
- Schreiter Lab (17) Apply Schreiter Lab filter
- Sgro Lab (20) Apply Sgro Lab filter
- Simpson Lab (5) Apply Simpson Lab filter
- Singer Lab (43) Apply Singer Lab filter
- Spruston Lab (36) Apply Spruston Lab filter
- Stern Lab (83) Apply Stern Lab filter
- Sternson Lab (7) Apply Sternson Lab filter
- Stringer Lab (3) Apply Stringer Lab filter
- Svoboda Lab (4) Apply Svoboda Lab filter
- Tebo Lab (24) Apply Tebo Lab filter
- Tillberg Lab (3) Apply Tillberg Lab filter
- Tjian Lab (47) Apply Tjian Lab filter
- Truman Lab (30) Apply Truman Lab filter
- Turaga Lab (12) Apply Turaga Lab filter
- Turner Lab (11) Apply Turner Lab filter
- Wang (Shaohe) Lab (19) Apply Wang (Shaohe) Lab filter
- Wu Lab (1) Apply Wu Lab filter
- Zlatic Lab (2) Apply Zlatic Lab filter
- Zuker Lab (20) Apply Zuker Lab filter
Associated Project Team
Publication Date
- 2023 (1) Apply 2023 filter
- 2022 (26) Apply 2022 filter
- 2021 (19) Apply 2021 filter
- 2020 (19) Apply 2020 filter
- 2019 (25) Apply 2019 filter
- 2018 (26) Apply 2018 filter
- 2017 (31) Apply 2017 filter
- 2016 (18) Apply 2016 filter
- 2015 (57) Apply 2015 filter
- 2014 (46) Apply 2014 filter
- 2013 (58) Apply 2013 filter
- 2012 (78) Apply 2012 filter
- 2011 (92) Apply 2011 filter
- 2010 (100) Apply 2010 filter
- 2009 (102) Apply 2009 filter
- 2008 (100) Apply 2008 filter
- 2007 (85) Apply 2007 filter
- 2006 (89) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
- Remove Non-Janelia filter Non-Janelia
1416 Publications
Showing 1321-1330 of 1416 resultsNuclear genes encoding mitochondrial proteins are regulated by carbon source with significant heterogeneity among four Saccharomyces cerevisiae strains. This strain-dependent variation is seen both in respiratory capacity of the cells and in the expression of beta-galactosidase reporter fusions to the promoters of CYB2, CYC1, CYC3, MnSOD, and RPO41.
Differentiation of the Drosophila retina is asynchronous: it starts at the posterior margin of the eye imaginal disc and progresses anteriorly over two days. During this time the disc continues to grow, increasing in size by approximately eightfold. An indentation in the epithelium, the morphogenetic furrow, marks the front edge of the differentiation wave. Anterior progression of the furrow is thought to be driven by signals emanating from differentiating photoreceptor cells in the posterior eye disc. A good candidate for such a signal is the product of the hedgehog (hh) gene; it is expressed, and presumably secreted, by differentiating photoreceptors and its function is required for continued furrow movement. Here we show that ectopic expression of hedgehog sets in motion ectopic furrows in the anterior eye disc. In addition to changes in cell shape, these ectopic furrows are associated with a tightly orchestrated series of events, including proliferation, cell cycle synchronization and pattern formation, that parallel normal furrow progression. We propose that the morphogenetic furrow coincides with a transient boundary that coordinates growth and differentiation of the eye disc, and that hedgehog is necessary and sufficient to propagate this boundary across the epithelium.
We can resolve multiple discrete features within a focal region of m spatial dimensions by first isolating each on the basis of n >/= 1 unique optical characteristics and then measuring their relative spatial coordinates. The minimum acceptable separation between features depends on the point-spread function in the (m + n)d-dimensional space formed by the spatial coordinates and the optical parameters, whereas the absolute spatial resolution is determined by the accuracy to which the coordinates can be measured. Estimates of each suggest that near-field fluorescence excitation microscopy/spectroscopy with molecular sensitivity and spatial resolution is possible.
Commentary: Inspired by my earlier work (see below) in single molecule imaging and the isolation of multiple exciton recombination sites within a single probe volume, here I proposed the principle which would eventually lead to PALM. Indeed, all methods of localization microscopy, including PALM, fPALM, PALMIRA, STORM, dSTORM, PAINT, GSDIM, etc. are specific embodiments of the general principle of single molecule isolation and localization I introduced here.
We describe the life cycle and general biology of the tropical cerataphidine aphid Cerataphis fransseni. We demonstrate that this aphid migrates between trees of Styrax benzoin and various species of palms; palm-feeding populations have previously been known as C. variabilis and C. palmae, which now become synonyms of C. fransseni. On S. benzoin the fundatrix induces a relatively simple gall which can contain >6000 aphids at maturity with a large number of reproductively sterile soldiers that protect the gall from predators. These galls are apparently produced throughout the year. Colonies on the secondary host plants, palms, are apparently obligately tended by ants whereas colonies within galls on Styrax are never tended by ants. We discuss the life cycle of this tropical aphid with respect to hypotheses for the evolution and maintenance of host alternation.
1. Properties of dendritic glutamate receptor (GluR) channels were investigated using fast application of glutamate to outside-out membrane patches isolated from the apical dendrites of CA3 and CA1 pyramidal neurons in rat hippocampal slices. CA3 patches were formed (15-76 microns from the soma) in the region of mossy fibre (MF) synapses, and CA1 patches (25-174 microns from the soma) in the region of Schaffer collateral (SC) innervation. 2. Dual-component responses consisting of a rapidly rising and decaying component followed by a second, substantially slower, component were elicited by 1 ms pulses of 1 mM glutamate in the presence of 10 microM glycine and absence of external Mg2+. The fast component was selectively blocked by 2-5 microM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) and the slow component by 30 microM D-2-amino-5-phosphonopentanoic acid (D-AP5), suggesting that the fast and slow components were mediated by the GluR channels of the L-alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) and NMDA type, respectively. The peak amplitude ratio of the NMDA to AMPA receptor-mediated components varied between 0.03 and 0.62 in patches from both CA3 and CA1 dendrites. Patches lacking either component were rarely observed. 3. The peak current-voltage (I-V) relationship of the fast component was almost linear, whereas the I-V relationship of the slow component showed a region of negative slope in the presence of 1 mM external Mg2+. The reversal potential for both components was close to 0 mV. 4. Kainate-preferring GluR channels did not contribute appreciably to the response to glutamate. The responses to 100 ms pulses of 1 mM glutamate were mimicked by application of 1 mM AMPA, whereas 1 mM kainate produced much smaller, weakly desensitizing currents. This suggests that the fast component is primarily mediated by the action of glutamate on AMPA-preferring receptors. 5. The mean elementary conductance of AMPA receptor channels was about 10 pS, as estimated by non-stationary fluctuation analysis. The permeability of these channels to Ca2+ was low (approximately 5% of the permeability to Cs+). 6. The elementary conductance of NMDA receptor channels was larger, with a main conductance state of about 45 pS. These channels were 3.6 times more permeable to Ca2+ than to Cs+.(ABSTRACT TRUNCATED AT 400 WORDS)
Critical features of the mitochondrial leading-strand DNA replication origin are conserved from Saccharomyces cerevisiae to humans. These include a promoter and a downstream GC-rich sequence block (CSBII) that encodes rGs within the primer RNA. During in vitro transcription at yeast mitochondrial replication origins, there is stable and persistent RNA-DNA hybrid formation that begins at the 5’ end of the rG region. The short rG-dC sequence is the necessary and sufficient nucleic acid element for establishing stable hybrids, and the presence of rGs within the RNA strand of the RNA-DNA hybrid is required. The efficiency of hybrid formation depends on the length of RNA synthesized 5’ to CSBII and the type of RNA polymerase employed. Once made, the RNA strand of an RNA-DNA hybrid can serve as an effective primer for mitochondrial DNA polymerase. These results reveal a new mechanism for persistent RNA-DNA hybrid formation and suggest a step in priming mitochondrial DNA replication that requires both mitochondrial RNA polymerase and an rG-dC sequence-specific event to form an extensive RNA-DNA hybrid.
The gall-forming aphidCerataphis fransseni produces soldiers that defend against predators. Soldiers are produced soon after colony foundation and the number of soldiers increases nonlinearly during colony growth. The number of soldiers scales to the square-root of the number of non-soldiers and linearly to the surface area of the gall. This suggests that soldiers are produced to defend an area, for example the perimeter of the colony or the surface of the gall, rather than individual aphids.
In insects, the neuropeptide eclosion hormone (EH) acts on the CNS to evoke the stereotyped behaviors that cause ecdysis, the shedding of the cuticle at the end of each molt. Concomitantly, EH induces an increase in cyclic GMP (cGMP). Using antibodies against this second messenger, we show that this increase is confined to a network of 50 peptidergic neurons distributed throughout the CNS. Increases appeared 30 min after EH treatment, spread rapidly throughout these neurons, and were extremely long lived. We show that this response is synaptically driven, and does not involve the soluble, nitric oxide (NO)-activated, guanylate cyclase. Stereotyped variations in the duration of the cGMP response among neurons suggest a role in coordinating responses having different latencies and durations.
Given a relatively short query stringW of lengthP, a long subject stringA of lengthN, and a thresholdD, theapproximate keyword search problem is to find all substrings ofA that align withW with not more than D insertions, deletions, and mismatches. In typical applications, such as searching a DNA sequence database, the size of the “database”A is much larger than that of the queryW, e.g.,N is on the order of millions or billions andP is a hundred to a thousand. In this paper we present an algorithm that given a precomputedindex of the databaseA, finds rare matches in time that issublinear inN, i.e.,N c for somec<1. The sequenceA must be overa. finite alphabet σ. More precisely, our algorithm requires 0(DN pow(ɛ) logN) expected-time where ɛ=D/P is the maximum number of differences as a percentage of query length, and pow(ɛ) is an increasing and concave function that is 0 when ɛ=0. Thus the algorithm is superior to current O(DN) algorithms when ɛ is small enough to guarantee that pow(ɛ) < 1. As seen in the paper, this is true for a wide range of ɛ, e.g., ɛ. up to 33% for DNA sequences (¦⌆¦=4) and 56% for proteins sequences (¦⌆¦=20). In preliminary practical experiments, the approach gives a 50-to 500-fold improvement over previous algorithms for prolems of interest in molecular biology.
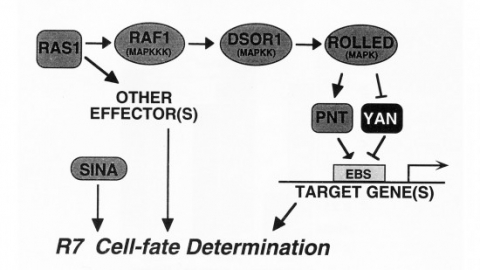
We show that the activities of two Ets-related transcription factors required for normal eye development in Drosophila, pointed and yan, are regulated by the Ras1/MAPK pathway. The pointed gene codes for two related proteins, and we show that one form is a constitutive activator of transcription, while the activity of the other form is stimulated by the Ras1/MAPK pathway. Mutation of the single consensus MAPK phosphorylation site in the second form abrogates this responsiveness. yan is a negative regulator of photoreceptor determination, and genetic data suggest that it acts as an antagonist of Ras1. We demonstrate that yan can repress transcription and that this repression activity is negatively regulated by the Ras1/MAPK signal, most likely through direct phosphorylation of yan by MAPK.