Filter
Associated Lab
- Aso Lab (30) Apply Aso Lab filter
- Betzig Lab (1) Apply Betzig Lab filter
- Bock Lab (2) Apply Bock Lab filter
- Branson Lab (9) Apply Branson Lab filter
- Card Lab (5) Apply Card Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Dickson Lab (2) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Funke Lab (2) Apply Funke Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (2) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Hess Lab (6) Apply Hess Lab filter
- Jayaraman Lab (7) Apply Jayaraman Lab filter
- Lippincott-Schwartz Lab (1) Apply Lippincott-Schwartz Lab filter
- Looger Lab (2) Apply Looger Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Reiser Lab (16) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Romani Lab (2) Apply Romani Lab filter
- Rubin Lab (147) Apply Rubin Lab filter
- Saalfeld Lab (5) Apply Saalfeld Lab filter
- Scheffer Lab (8) Apply Scheffer Lab filter
- Schreiter Lab (1) Apply Schreiter Lab filter
- Simpson Lab (3) Apply Simpson Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Stern Lab (1) Apply Stern Lab filter
- Svoboda Lab (3) Apply Svoboda Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (4) Apply Truman Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
- Turner Lab (5) Apply Turner Lab filter
- Zuker Lab (1) Apply Zuker Lab filter
Associated Project Team
- CellMap (1) Apply CellMap filter
- Fly Functional Connectome (4) Apply Fly Functional Connectome filter
- Fly Olympiad (3) Apply Fly Olympiad filter
- FlyEM (12) Apply FlyEM filter
- FlyLight (20) Apply FlyLight filter
- GENIE (1) Apply GENIE filter
- Transcription Imaging (1) Apply Transcription Imaging filter
Publication Date
- 2025 (8) Apply 2025 filter
- 2024 (4) Apply 2024 filter
- 2023 (5) Apply 2023 filter
- 2022 (1) Apply 2022 filter
- 2021 (4) Apply 2021 filter
- 2020 (9) Apply 2020 filter
- 2019 (6) Apply 2019 filter
- 2018 (7) Apply 2018 filter
- 2017 (15) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (16) Apply 2015 filter
- 2014 (9) Apply 2014 filter
- 2013 (5) Apply 2013 filter
- 2012 (8) Apply 2012 filter
- 2011 (4) Apply 2011 filter
- 2010 (4) Apply 2010 filter
- 2009 (2) Apply 2009 filter
- 2008 (4) Apply 2008 filter
- 2007 (2) Apply 2007 filter
- 2006 (1) Apply 2006 filter
- 2002 (1) Apply 2002 filter
- 2000 (2) Apply 2000 filter
- 1999 (1) Apply 1999 filter
- 1997 (1) Apply 1997 filter
- 1995 (2) Apply 1995 filter
- 1994 (2) Apply 1994 filter
- 1993 (2) Apply 1993 filter
- 1992 (1) Apply 1992 filter
- 1991 (2) Apply 1991 filter
- 1990 (3) Apply 1990 filter
- 1989 (2) Apply 1989 filter
- 1987 (2) Apply 1987 filter
- 1986 (1) Apply 1986 filter
- 1985 (1) Apply 1985 filter
- 1984 (1) Apply 1984 filter
- 1983 (1) Apply 1983 filter
- 1982 (2) Apply 1982 filter
- 1981 (1) Apply 1981 filter
- 1979 (1) Apply 1979 filter
- 1973 (1) Apply 1973 filter
Type of Publication
147 Publications
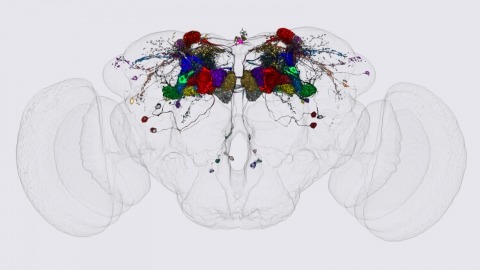
Showing 131-140 of 147 resultsWe identified the neurons comprising the Drosophila mushroom body (MB), an associative center in invertebrate brains, and provide a comprehensive map describing their potential connections. Each of the 21 MB output neuron (MBON) types elaborates segregated dendritic arbors along the parallel axons of ∼2000 Kenyon cells, forming 15 compartments that collectively tile the MB lobes. MBON axons project to five discrete neuropils outside of the MB and three MBON types form a feedforward network in the lobes. Each of the 20 dopaminergic neuron (DAN) types projects axons to one, or at most two, of the MBON compartments. Convergence of DAN axons on compartmentalized Kenyon cell-MBON synapses creates a highly ordered unit that can support learning to impose valence on sensory representations. The elucidation of the complement of neurons of the MB provides a comprehensive anatomical substrate from which one can infer a functional logic of associative olfactory learning and memory.
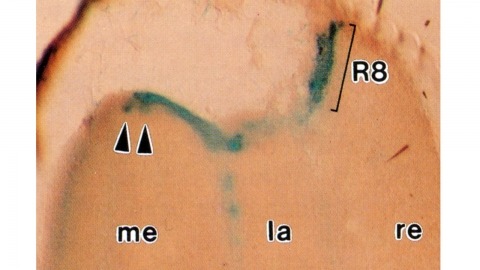
Histological staining of wild-type and sevenless transgenic Drosophila melanogaster bearing Rh3-lacZ fusion genes permits the selective visualization of polarization-sensitive R7 and R8 photoreceptor cells located along the dorsal anterior eye margin. Diffusion of beta-galactosidase throughout these cells reveals that they project long axons to the two most peripheral synaptic target rows of the dorsal posterior medulla, defining a specialized marginal zone of this optic lobe. Comparison of the staining patterns of marginal and nonmarginal Rh3-lacZ-expressing photoreceptor cells in the same histological preparations suggest that the marginal cells possess morphologically specialized axons and synaptic terminals. These findings are discussed with reference to the neuroanatomy of the corresponding dorsal marginal eye and optic lobe regions of the larger dipterans Musca and Calliphora, and in relation to the ability of Drosophila to orient to polarized light.
Drosophila melanogaster plays an important role in molecular, genetic, and genomic studies of heredity, development, metabolism, behavior, and human disease. The initial reference genome sequence reported more than a decade ago had a profound impact on progress in Drosophila research, and improving the accuracy and completeness of this sequence continues to be important to further progress. We previously described improvement of the 117-Mb sequence in the euchromatic portion of the genome and 21 Mb in the heterochromatic portion, using a whole-genome shotgun assembly, BAC physical mapping, and clone-based finishing. Here, we report an improved reference sequence of the single-copy and middle-repetitive regions of the genome, produced using cytogenetic mapping to mitotic and polytene chromosomes, clone-based finishing and BAC fingerprint verification, ordering of scaffolds by alignment to cDNA sequences, incorporation of other map and sequence data, and validation by whole-genome optical restriction mapping. These data substantially improve the accuracy and completeness of the reference sequence and the order and orientation of sequence scaffolds into chromosome arm assemblies. Representation of the Y chromosome and other heterochromatic regions is particularly improved. The new 143.9-Mb reference sequence, designated Release 6, effectively exhausts clone-based technologies for mapping and sequencing. Highly repeat-rich regions, including large satellite blocks and functional elements such as the ribosomal RNA genes and the centromeres, are largely inaccessible to current sequencing and assembly methods and remain poorly represented. Further significant improvements will require sequencing technologies that do not depend on molecular cloning and that produce very long reads.
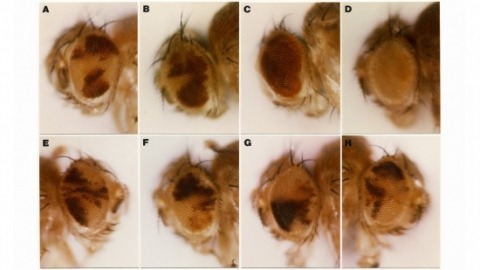
Development of the Drosophila retina occurs asynchronously; differentiation, its front marked by the morphogenetic furrow, progresses across the eye disc epithelium over a 2 day period. We have investigated the mechanism by which this front advances, and our results suggest that developing retinal cells drive the progression of morphogenesis utilizing the products of the hedgehog (hh) and decapentaplegic (dpp) genes. Analysis of hh and dpp genetic mosaics indicates that the products of these genes act as diffusible signals in this process. Expression of dpp in the morphogenetic furrow is closely correlated with the progression of the furrow under a variety of conditions. We show that hh, synthesized by differentiating cells, induces the expression of dpp, which appears to be a primary mediator of furrow movement.
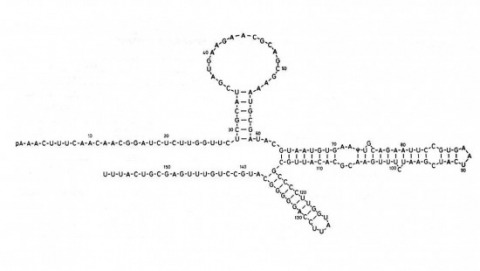
We show that the germline specificity of P element transposition is controlled at the level of mRNA splicing and not at the level of transcription. In the major P element RNA transcript, isolated from somatic cells, the first three open reading frames are joined by the removal of two introns. Using in vitro mutagenesis and genetic analysis we demonstrate the existence of a third intron whose removal is required for transposase production. We propose that this intron is only removed in the germline and that its removal is the sole basis for the germline restriction of P element transposition.
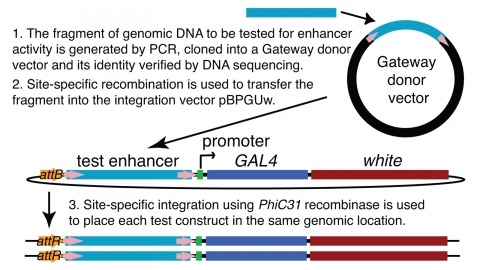
We demonstrate the feasibility of generating thousands of transgenic Drosophila melanogaster lines in which the expression of an exogenous gene is reproducibly directed to distinct small subsets of cells in the adult brain. We expect the expression patterns produced by the collection of 5,000 lines that we are currently generating to encompass all neurons in the brain in a variety of intersecting patterns. Overlapping 3-kb DNA fragments from the flanking noncoding and intronic regions of genes thought to have patterned expression in the adult brain were inserted into a defined genomic location by site-specific recombination. These fragments were then assayed for their ability to function as transcriptional enhancers in conjunction with a synthetic core promoter designed to work with a wide variety of enhancer types. An analysis of 44 fragments from four genes found that >80% drive expression patterns in the brain; the observed patterns were, on average, comprised of <100 cells. Our results suggest that the D. melanogaster genome contains >50,000 enhancers and that multiple enhancers drive distinct subsets of expression of a gene in each tissue and developmental stage. We expect that these lines will be valuable tools for neuroanatomy as well as for the elucidation of neuronal circuits and information flow in the fly brain.
The Mushroom Body (MB) is the primary location of stored associative memories in the Drosophila brain. We discuss recent advances in understanding the MB's neuronal circuits made using advanced light microscopic methods and cell-type-specific genetic tools. We also review how the compartmentalized nature of the MB's organization allows this brain area to form and store memories with widely different dynamics.
The stability of elements of three different dispersed repeated gene families in the genome of Drosophila tissue culture cells has been examined. Different amounts of sequences homologous to elements of 412, copia and 297 dispersed repeated gene families are found in the genomes of D. melanogaster embryonic and tissue culture cells. In general the amount of these sequences is increased in the cell lines. The additional sequences homologous to 412, copia and 297 occur as intact elements and are dispersed to new sites in the cell culture genome. It appears that these elements can insert at many alternative sites. We also describe a DNA sequence arrangement found in the D. melanogaster embryo genome which appears to result from a transposition of an element of the copia dispersed repeated gene family into a new chromosomal site. The mechanism of insertion of this copia element is precise to within 90 bp and may involve a region of weak sequence homology between the site of insertion and the direct terminal repeats of the copia element.
Nervous systems combine lower-level sensory signals to detect higher-order stimulus features critical to survival, such as the visual looming motion created by an imminent collision or approaching predator. Looming-sensitive neurons have been identified in diverse animal species. Different large-scale visual features such as looming often share local cues, which means loom-detecting neurons face the challenge of rejecting confounding stimuli. Here we report the discovery of an ultra-selective looming detecting neuron, lobula plate/lobula columnar, type II (LPLC2) in Drosophila, and show how its selectivity is established by radial motion opponency. In the fly visual system, directionally selective small-field neurons called T4 and T5 form a spatial map in the lobula plate, where they each terminate in one of four retinotopic layers, such that each layer responds to motion in a different cardinal direction. Single-cell anatomical analysis reveals that each arm of the LPLC2 cross-shaped primary dendrites ramifies in one of these layers and extends along that layer's preferred motion direction. In vivo calcium imaging demonstrates that, as their shape predicts, individual LPLC2 neurons respond strongly to outward motion emanating from the centre of the neuron's receptive field. Each dendritic arm also receives local inhibitory inputs directionally selective for inward motion opposing the excitation. This radial motion opponency generates a balance of excitation and inhibition that makes LPLC2 non-responsive to related patterns of motion such as contraction, wide-field rotation or luminance change. As a population, LPLC2 neurons densely cover visual space and terminate onto the giant fibre descending neurons, which drive the jump muscle motor neuron to trigger an escape take off. Our findings provide a mechanistic description of the selective feature detection that flies use to discern and escape looming threats.