Filter
Associated Lab
- Aguilera Castrejon Lab (1) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (45) Apply Ahrens Lab filter
- Aso Lab (39) Apply Aso Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (98) Apply Betzig Lab filter
- Beyene Lab (4) Apply Beyene Lab filter
- Bock Lab (14) Apply Bock Lab filter
- Branson Lab (45) Apply Branson Lab filter
- Card Lab (34) Apply Card Lab filter
- Cardona Lab (44) Apply Cardona Lab filter
- Chklovskii Lab (10) Apply Chklovskii Lab filter
- Clapham Lab (11) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (8) Apply Darshan Lab filter
- Dickson Lab (32) Apply Dickson Lab filter
- Druckmann Lab (21) Apply Druckmann Lab filter
- Dudman Lab (34) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (4) Apply Egnor Lab filter
- Espinosa Medina Lab (12) Apply Espinosa Medina Lab filter
- Feliciano Lab (6) Apply Feliciano Lab filter
- Fetter Lab (31) Apply Fetter Lab filter
- Fitzgerald Lab (15) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (34) Apply Funke Lab filter
- Gonen Lab (59) Apply Gonen Lab filter
- Grigorieff Lab (34) Apply Grigorieff Lab filter
- Harris Lab (48) Apply Harris Lab filter
- Heberlein Lab (13) Apply Heberlein Lab filter
- Hermundstad Lab (17) Apply Hermundstad Lab filter
- Hess Lab (68) Apply Hess Lab filter
- Ilanges Lab (1) Apply Ilanges Lab filter
- Jayaraman Lab (39) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (1) Apply Johnson Lab filter
- Karpova Lab (13) Apply Karpova Lab filter
- Keleman Lab (8) Apply Keleman Lab filter
- Keller Lab (60) Apply Keller Lab filter
- Lavis Lab (123) Apply Lavis Lab filter
- Lee (Albert) Lab (29) Apply Lee (Albert) Lab filter
- Leonardo Lab (19) Apply Leonardo Lab filter
- Li Lab (1) Apply Li Lab filter
- Lippincott-Schwartz Lab (89) Apply Lippincott-Schwartz Lab filter
- Liu (Zhe) Lab (53) Apply Liu (Zhe) Lab filter
- Looger Lab (136) Apply Looger Lab filter
- Magee Lab (31) Apply Magee Lab filter
- Menon Lab (12) Apply Menon Lab filter
- Murphy Lab (6) Apply Murphy Lab filter
- O'Shea Lab (3) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Pachitariu Lab (28) Apply Pachitariu Lab filter
- Pastalkova Lab (5) Apply Pastalkova Lab filter
- Pavlopoulos Lab (7) Apply Pavlopoulos Lab filter
- Pedram Lab (3) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (43) Apply Reiser Lab filter
- Riddiford Lab (20) Apply Riddiford Lab filter
- Romani Lab (28) Apply Romani Lab filter
- Rubin Lab (101) Apply Rubin Lab filter
- Saalfeld Lab (43) Apply Saalfeld Lab filter
- Satou Lab (1) Apply Satou Lab filter
- Scheffer Lab (36) Apply Scheffer Lab filter
- Schreiter Lab (44) Apply Schreiter Lab filter
- Shroff Lab (22) Apply Shroff Lab filter
- Simpson Lab (18) Apply Simpson Lab filter
- Singer Lab (37) Apply Singer Lab filter
- Spruston Lab (55) Apply Spruston Lab filter
- Stern Lab (69) Apply Stern Lab filter
- Sternson Lab (47) Apply Sternson Lab filter
- Stringer Lab (25) Apply Stringer Lab filter
- Svoboda Lab (131) Apply Svoboda Lab filter
- Tebo Lab (7) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (14) Apply Tillberg Lab filter
- Tjian Lab (17) Apply Tjian Lab filter
- Truman Lab (58) Apply Truman Lab filter
- Turaga Lab (34) Apply Turaga Lab filter
- Turner Lab (24) Apply Turner Lab filter
- Vale Lab (6) Apply Vale Lab filter
- Voigts Lab (1) Apply Voigts Lab filter
- Wang (Meng) Lab (9) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (5) Apply Wang (Shaohe) Lab filter
- Wu Lab (8) Apply Wu Lab filter
- Zlatic Lab (26) Apply Zlatic Lab filter
- Zuker Lab (5) Apply Zuker Lab filter
Associated Project Team
- CellMap (4) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- Fly Descending Interneuron (10) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (51) Apply FlyEM filter
- FlyLight (46) Apply FlyLight filter
- GENIE (40) Apply GENIE filter
- Integrative Imaging (1) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (16) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (24) Apply Tool Translation Team (T3) filter
- Transcription Imaging (45) Apply Transcription Imaging filter
Associated Support Team
- Project Pipeline Support (1) Apply Project Pipeline Support filter
- Anatomy and Histology (18) Apply Anatomy and Histology filter
- Cryo-Electron Microscopy (33) Apply Cryo-Electron Microscopy filter
- Electron Microscopy (12) Apply Electron Microscopy filter
- Fly Facility (39) Apply Fly Facility filter
- Gene Targeting and Transgenics (11) Apply Gene Targeting and Transgenics filter
- Integrative Imaging (10) Apply Integrative Imaging filter
- Janelia Experimental Technology (35) Apply Janelia Experimental Technology filter
- Management Team (1) Apply Management Team filter
- Molecular Genomics (15) Apply Molecular Genomics filter
- Primary & iPS Cell Culture (13) Apply Primary & iPS Cell Culture filter
- Project Technical Resources (36) Apply Project Technical Resources filter
- Quantitative Genomics (19) Apply Quantitative Genomics filter
- Scientific Computing Software (59) Apply Scientific Computing Software filter
- Scientific Computing Systems (6) Apply Scientific Computing Systems filter
- Viral Tools (14) Apply Viral Tools filter
- Vivarium (6) Apply Vivarium filter
Publication Date
- 2024 (132) Apply 2024 filter
- 2023 (175) Apply 2023 filter
- 2022 (166) Apply 2022 filter
- 2021 (174) Apply 2021 filter
- 2020 (177) Apply 2020 filter
- 2019 (177) Apply 2019 filter
- 2018 (206) Apply 2018 filter
- 2017 (186) Apply 2017 filter
- 2016 (191) Apply 2016 filter
- 2015 (195) Apply 2015 filter
- 2014 (190) Apply 2014 filter
- 2013 (136) Apply 2013 filter
- 2012 (112) Apply 2012 filter
- 2011 (98) Apply 2011 filter
- 2010 (61) Apply 2010 filter
- 2009 (56) Apply 2009 filter
- 2008 (40) Apply 2008 filter
- 2007 (21) Apply 2007 filter
- 2006 (3) Apply 2006 filter
2496 Janelia Publications
Showing 2411-2420 of 2496 resultsThe analysis of single particle trajectories plays an important role in elucidating dynamics within complex environments such as those found in living cells. However, the characterization of intracellular particle motion is often confounded by confinement of the particles within non-trivial subcellular geometries. Here, we focus specifically on the case of particles undergoing Brownian motion within a tubular network, as found in some cellular organelles. An unraveling algorithm is developed to uncouple particle motion from the confining network structure, allowing for an accurate extraction of the diffusion coefficient, as well as differentiating between Brownian and fractional Brownian dynamics. We validate the algorithm with simulated trajectories and then highlight its application to an example system: analyzing the motion of membrane proteins confined in the tubules of the peripheral endoplasmic reticulum in mammalian cells. We show that these proteins undergo diffusive motion with a well-characterized diffusivity. Our algorithm provides a generally applicable approach for disentangling geometric morphology and particle dynamics in networked architectures.
Segmentation of objects in microscopy images is required for many biomedical applications. We introduce object-centric embeddings (OCEs), which embed image patches such that the spatial offsets between patches cropped from the same object are preserved. Those learnt embeddings can be used to delineate individual objects and thus obtain instance segmentations. Here, we show theoretically that, under assumptions commonly found in microscopy images, OCEs can be learnt through a self-supervised task that predicts the spatial offset between image patches. Together, this forms an unsupervised cell instance segmentation method which we evaluate on nine diverse large-scale microscopy datasets. Segmentations obtained with our method lead to substantially improved results, compared to state-of-the-art baselines on six out of nine datasets, and perform on par on the remaining three datasets. If ground-truth annotations are available, our method serves as an excellent starting point for supervised training, reducing the required amount of ground-truth needed by one order of magnitude, thus substantially increasing the practical applicability of our method. Source code is available at github.com/funkelab/cellulus.
Background: Segmenting electron microscopy (EM) images of cellular and subcellular processes in the nervous system is a key step in many bioimaging pipelines involving classification and labeling of ultrastructures. However, fully automated techniques to segment images are often susceptible to noise and heterogeneity in EM images (e.g. different histological preparations, different organisms, different brain regions, etc.). Supervised techniques to address this problem are often helpful but require large sets of training data, which are often difficult to obtain in practice, especially across many conditions. Results: We propose a new, principled unsupervised algorithm to segment EM images using a two-step approach: edge detection via salient watersheds following by robust region merging. We performed experiments to gather EM neuroimages of two organisms (mouse and fruit fly) using different histological preparations and generated manually curated ground-truth segmentations. We compared our algorithm against several state-of- the-art unsupervised segmentation algorithms and found superior performance using two standard measures of under-and over-segmentation error. Conclusions: Our algorithm is general and may be applicable to other large-scale segmentation problems for bioimages.
Neuroendocrine systems in animals maintain organismal homeostasis and regulate stress response. Although a great deal of work has been done on the neuropeptides and hormones that are released and act on target organs in the periphery, the synaptic inputs onto these neuroendocrine outputs in the brain are less well understood. Here, we use the transmission electron microscopy reconstruction of a whole central nervous system in the larva to elucidate the sensory pathways and the interneurons that provide synaptic input to the neurosecretory cells projecting to the endocrine organs. Predicted by network modeling, we also identify a new carbon dioxide-responsive network that acts on a specific set of neurosecretory cells and that includes those expressing corazonin (Crz) and diuretic hormone 44 (Dh44) neuropeptides. Our analysis reveals a neuronal network architecture for combinatorial action based on sensory and interneuronal pathways that converge onto distinct combinations of neuroendocrine outputs.
Phylogenetic footprinting has revealed that cis-regulatory enhancers consist of conserved DNA sequence clusters (CSCs). Currently, there is no systematic approach for enhancer discovery and analysis that takes full-advantage of the sequence information within enhancer CSCs.
Reconstructing neuron morphologies from fluorescence microscope images plays a critical role in neuroscience studies. It relies on image segmentation to produce initial masks either for further processing or final results to represent neuronal morphologies. This has been a challenging step due to the variation and complexity of noisy intensity patterns in neuron images acquired from microscopes. Whereas progresses in deep learning have brought the goal of accurate segmentation much closer to reality, creating training data for producing powerful neural networks is often laborious. To overcome the difficulty of obtaining a vast number of annotated data, we propose a novel strategy of using two-stage generative models to simulate training data with voxel-level labels. Trained upon unlabeled data by optimizing a novel objective function of preserving predefined labels, the models are able to synthesize realistic 3D images with underlying voxel labels. We showed that these synthetic images could train segmentation networks to obtain even better performance than manually labeled data. To demonstrate an immediate impact of our work, we further showed that segmentation results produced by networks trained upon synthetic data could be used to improve existing neuron reconstruction methods.
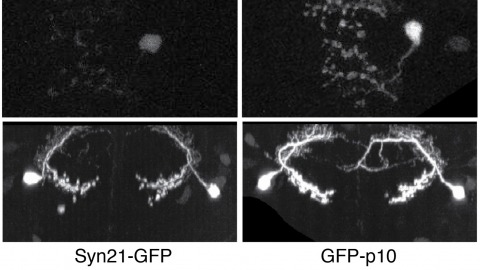
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibire(ts1), and for the use of cytoplasmic GFP to image the fine processes of neurons.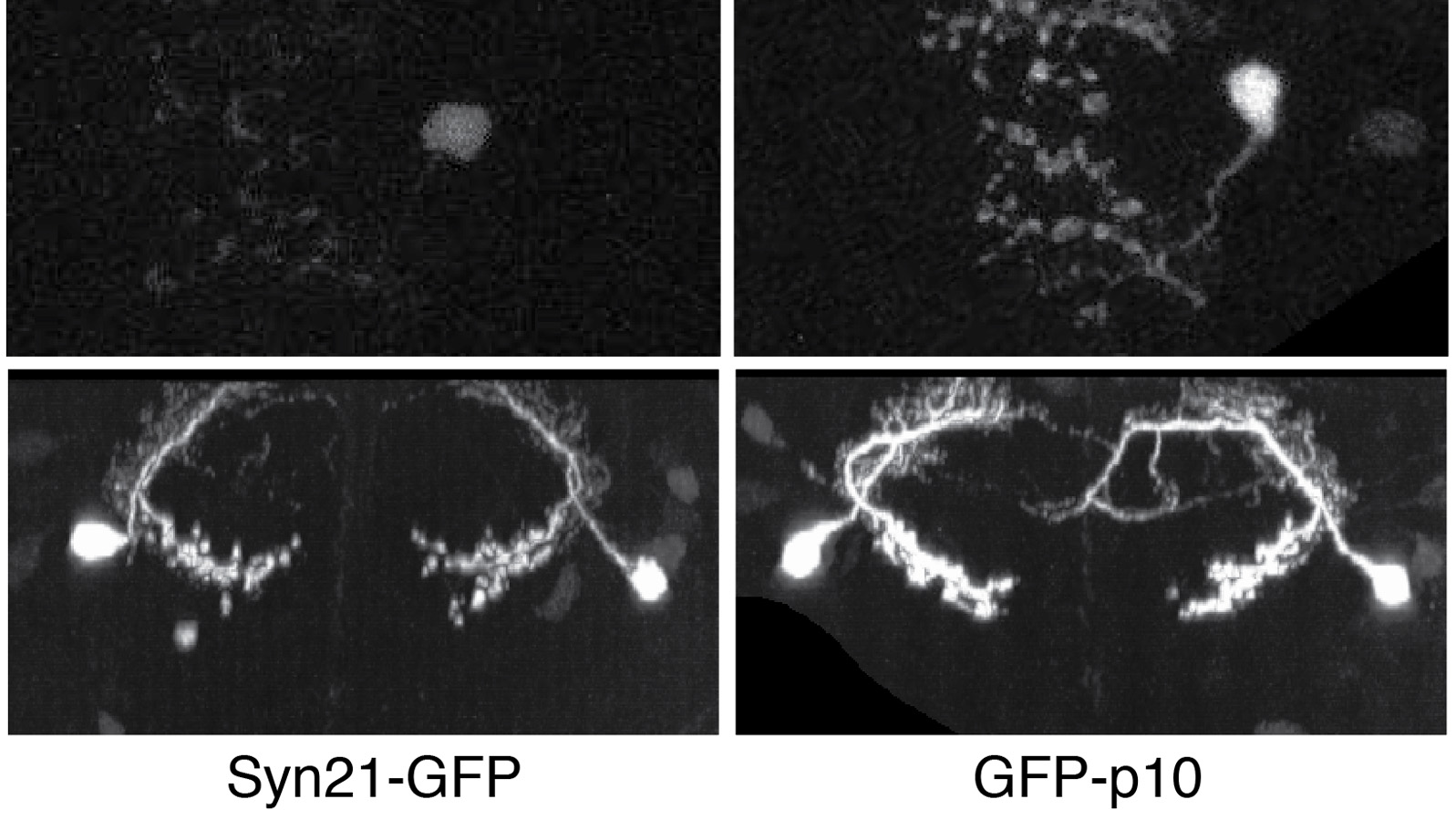
Capping Protein (CP) plays a central role in the creation of the Arp2/3-generated branched actin networks comprising lamellipodia and pseudopodia by virtue of its ability to cap the actin filament barbed end, which promotes Arp2/3-dependent filament nucleation and optimal branching. The highly conserved protein V-1/Myotrophin binds CP tightly in vitro to render it incapable of binding the barbed end. Here we addressed the physiological significance of this CP antagonist in Dictyostelium, which expresses a V-1 homolog that we show is very similar biochemically to mouse V-1. Consistent with previous studies of CP knockdown, overexpression of V-1 in Dictyostelium reduced the size of pseudopodia and the cortical content of Arp2/3 and induced the formation of filopodia. Importantly, these effects scaled positively with the degree of V-1 overexpression and were not seen with a V-1 mutant that cannot bind CP. V-1 is present in molar excess over CP, suggesting that it suppresses CP activity in the cytoplasm at steady state. Consistently, cells devoid of V-1, like cells overexpressing CP described previously, exhibited a significant decrease in cellular F-actin content. Moreover, V-1-null cells exhibited pronounced defects in macropinocytosis and chemotactic aggregation that were rescued by V-1, but not by the V-1 mutant. Together, these observations demonstrate that V-1 exerts significant influence in vivo on major actin-based processes via its ability to sequester CP. Finally, we present evidence that V-1's ability to sequester CP is regulated by phosphorylation, suggesting that cells may manipulate the level of active CP to tune their "actin phenotype."