Filter
Associated Lab
- Cardona Lab (1) Apply Cardona Lab filter
- Chklovskii Lab (1) Apply Chklovskii Lab filter
- Eddy/Rivas Lab (1) Apply Eddy/Rivas Lab filter
- Freeman Lab (1) Apply Freeman Lab filter
- Harris Lab (1) Apply Harris Lab filter
- Lavis Lab (1) Apply Lavis Lab filter
- Magee Lab (1) Apply Magee Lab filter
- Rubin Lab (1) Apply Rubin Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Stern Lab (1) Apply Stern Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Zlatic Lab (1) Apply Zlatic Lab filter
- Zuker Lab (1) Apply Zuker Lab filter
Associated Project Team
Associated Support Team
Publication Date
- January 28, 2015 (1) Apply January 28, 2015 filter
- January 27, 2015 (1) Apply January 27, 2015 filter
- January 21, 2015 (2) Apply January 21, 2015 filter
- January 19, 2015 (1) Apply January 19, 2015 filter
- January 16, 2015 (1) Apply January 16, 2015 filter
- January 15, 2015 (1) Apply January 15, 2015 filter
- January 13, 2015 (2) Apply January 13, 2015 filter
- January 7, 2015 (1) Apply January 7, 2015 filter
- Remove January 2015 filter January 2015
- Remove 2015 filter 2015
10 Janelia Publications
Showing 1-10 of 10 resultsThe Rfam database (available at http://rfam.xfam.org) is a collection of non-coding RNA families represented by manually curated sequence alignments, consensus secondary structures and annotation gathered from corresponding Wikipedia, taxonomy and ontology resources. In this article, we detail updates and improvements to the Rfam data and website for the Rfam 12.0 release. We describe the upgrade of our search pipeline to use Infernal 1.1 and demonstrate its improved homology detection ability by comparison with the previous version. The new pipeline is easier for users to apply to their own data sets, and we illustrate its ability to annotate RNAs in genomic and metagenomic data sets of various sizes. Rfam has been expanded to include 260 new families, including the well-studied large subunit ribosomal RNA family, and for the first time includes information on short sequence- and structure-based RNA motifs present within families.
Although undulatory swimming is observed in many organisms, the neuromuscular basis for undulatory movement patterns is not well understood. To better understand the basis for the generation of these movement patterns, we studied muscle activity in the nematode Caenorhabditis elegans. Caenorhabditis elegans exhibits a range of locomotion patterns: in low viscosity fluids the undulation has a wavelength longer than the body and propagates rapidly, while in high viscosity fluids or on agar media the undulatory waves are shorter and slower. Theoretical treatment of observed behaviour has suggested a large change in force-posture relationships at different viscosities, but analysis of bend propagation suggests that short-range proprioceptive feedback is used to control and generate body bends. How muscles could be activated in a way consistent with both these results is unclear. We therefore combined automated worm tracking with calcium imaging to determine muscle activation strategy in a variety of external substrates. Remarkably, we observed that across locomotion patterns spanning a threefold change in wavelength, peak muscle activation occurs approximately 45° (1/8th of a cycle) ahead of peak midline curvature. Although the location of peak force is predicted to vary widely, the activation pattern is consistent with required force in a model incorporating putative length- and velocity-dependence of muscle strength. Furthermore, a linear combination of local curvature and velocity can match the pattern of activation. This suggests that proprioception can enable the worm to swim effectively while working within the limitations of muscle biomechanics and neural control.
The apical tuft is the most remote area of the dendritic tree of neocortical pyramidal neurons. Despite its distal location, the apical dendritic tuft of layer 5 pyramidal neurons receives substantial excitatory synaptic drive and actively processes corticocortical input during behavior. The properties of the voltage-activated ion channels that regulate synaptic integration in tuft dendrites have, however, not been thoroughly investigated. Here, we use electrophysiological and optical approaches to examine the subcellular distribution and function of hyperpolarization-activated cyclic nucleotide-gated nonselective cation (HCN) channels in rat layer 5B pyramidal neurons. Outside-out patch recordings demonstrated that the amplitude and properties of ensemble HCN channel activity were uniform in patches excised from distal apical dendritic trunk and tuft sites. Simultaneous apical dendritic tuft and trunk whole-cell current-clamp recordings revealed that the pharmacological blockade of HCN channels decreased voltage compartmentalization and enhanced the generation and spread of apical dendritic tuft and trunk regenerative activity. Furthermore, multisite two-photon glutamate uncaging demonstrated that HCN channels control the amplitude and duration of synaptically evoked regenerative activity in the distal apical dendritic tuft. In contrast, at proximal apical dendritic trunk and somatic recording sites, the blockade of HCN channels decreased excitability. Dynamic-clamp experiments revealed that these compartment-specific actions of HCN channels were heavily influenced by the local and distributed impact of the high density of HCN channels in the distal apical dendritic arbor. The properties and subcellular distribution pattern of HCN channels are therefore tuned to regulate the interaction between integration compartments in layer 5B pyramidal neurons.
Specific labeling of biomolecules with bright fluorophores is the keystone of fluorescence microscopy. Genetically encoded self-labeling tag proteins can be coupled to synthetic dyes inside living cells, resulting in brighter reporters than fluorescent proteins. Intracellular labeling using these techniques requires cell-permeable fluorescent ligands, however, limiting utility to a small number of classic fluorophores. Here we describe a simple structural modification that improves the brightness and photostability of dyes while preserving spectral properties and cell permeability. Inspired by molecular modeling, we replaced the N,N-dimethylamino substituents in tetramethylrhodamine with four-membered azetidine rings. This addition of two carbon atoms doubles the quantum efficiency and improves the photon yield of the dye in applications ranging from in vitro single-molecule measurements to super-resolution imaging. The novel substitution is generalizable, yielding a palette of chemical dyes with improved quantum efficiencies that spans the UV and visible range.
In animals, Hox transcription factors define regional identity in distinct anatomical domains. How Hox genes encode this specificity is a paradox, because different Hox proteins bind with high affinity in vitro to similar DNA sequences. Here, we demonstrate that the Hox protein Ultrabithorax (Ubx) in complex with its cofactor Extradenticle (Exd) bound specifically to clusters of very low affinity sites in enhancers of the shavenbaby gene of Drosophila. These low affinity sites conferred specificity for Ubx binding in vivo, but multiple clustered sites were required for robust expression when embryos developed in variable environments. Although most individual Ubx binding sites are not evolutionarily conserved, the overall enhancer architecture-clusters of low affinity binding sites-is maintained and required for enhancer function. Natural selection therefore works at the level of the enhancer, requiring a particular density of low affinity Ubx sites to confer both specific and robust expression.
The mammalian taste system is responsible for sensing and responding to the five basic taste qualities: sweet, sour, bitter, salty and umami. Previously, we showed that each taste is detected by dedicated taste receptor cells (TRCs) on the tongue and palate epithelium. To understand how TRCs transmit information to higher neural centres, we examined the tuning properties of large ensembles of neurons in the first neural station of the gustatory system. Here, we generated and characterized a collection of transgenic mice expressing a genetically encoded calcium indicator in central and peripheral neurons, and used a gradient refractive index microendoscope combined with high-resolution two-photon microscopy to image taste responses from ganglion neurons buried deep at the base of the brain. Our results reveal fine selectivity in the taste preference of ganglion neurons; demonstrate a strong match between TRCs in the tongue and the principal neural afferents relaying taste information to the brain; and expose the highly specific transfer of taste information between taste cells and the central nervous system.
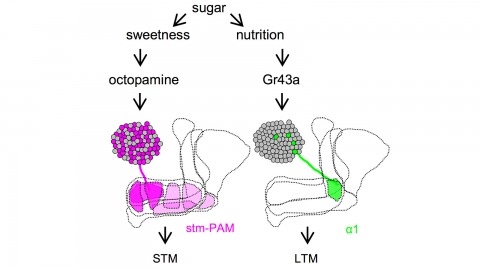
Drosophila melanogaster can acquire a stable appetitive olfactory memory when the presentation of a sugar reward and an odor are paired. However, the neuronal mechanisms by which a single training induces long-term memory are poorly understood. Here we show that two distinct subsets of dopamine neurons in the fly brain signal reward for short-term (STM) and long-term memories (LTM). One subset induces memory that decays within several hours, whereas the other induces memory that gradually develops after training. They convey reward signals to spatially segregated synaptic domains of the mushroom body (MB), a potential site for convergence. Furthermore, we identified a single type of dopamine neuron that conveys the reward signal to restricted subdomains of the mushroom body lobes and induces long-term memory. Constant appetitive memory retention after a single training session thus comprises two memory components triggered by distinct dopamine neurons.
Complex animal behaviors are built from dynamical relationships between sensory inputs, neuronal activity, and motor outputs in patterns with strategic value. Connecting these patterns illuminates how nervous systems compute behavior. Here, we study Drosophila larva navigation up temperature gradients toward preferred temperatures (positive thermotaxis). By tracking the movements of animals responding to fixed spatial temperature gradients or random temperature fluctuations, we calculate the sensitivity and dynamics of the conversion of thermosensory inputs into motor responses. We discover three thermosensory neurons in each dorsal organ ganglion (DOG) that are required for positive thermotaxis. Random optogenetic stimulation of the DOG thermosensory neurons evokes behavioral patterns that mimic the response to temperature variations. In vivo calcium and voltage imaging reveals that the DOG thermosensory neurons exhibit activity patterns with sensitivity and dynamics matched to the behavioral response. Temporal processing of temperature variations carried out by the DOG thermosensory neurons emerges in distinct motor responses during thermotaxis.
Tsetse flies (Glossina spp.), vectors of African trypanosomes, are distinguished by their specialized reproductive biology, defined by adenotrophic viviparity (maternal nourishment of progeny by glandular secretions followed by live birth). This trait has evolved infrequently among insects and requires unique reproductive mechanisms. A key event in Glossina reproduction involves the transition between periods of lactation and nonlactation (dry periods). Increased lipolysis, nutrient transfer to the milk gland, and milk-specific protein production characterize lactation, which terminates at the birth of the progeny and is followed by a period of involution. The dry stage coincides with embryogenesis of the progeny, during which lipid reserves accumulate in preparation for the next round of lactation. The obligate bacterial symbiont Wigglesworthia glossinidia is critical to tsetse reproduction and likely provides B vitamins required for metabolic processes underlying lactation and/or progeny development. Here we describe findings that utilized transcriptomics, physiological assays, and RNA interference-based functional analysis to understand different components of adenotrophic viviparity in tsetse flies.