Filter
Associated Lab
- Aso Lab (30) Apply Aso Lab filter
- Betzig Lab (1) Apply Betzig Lab filter
- Bock Lab (2) Apply Bock Lab filter
- Branson Lab (8) Apply Branson Lab filter
- Card Lab (5) Apply Card Lab filter
- Clapham Lab (1) Apply Clapham Lab filter
- Dickson Lab (2) Apply Dickson Lab filter
- Druckmann Lab (1) Apply Druckmann Lab filter
- Fetter Lab (1) Apply Fetter Lab filter
- Funke Lab (1) Apply Funke Lab filter
- Harris Lab (3) Apply Harris Lab filter
- Heberlein Lab (2) Apply Heberlein Lab filter
- Hermundstad Lab (2) Apply Hermundstad Lab filter
- Hess Lab (5) Apply Hess Lab filter
- Jayaraman Lab (6) Apply Jayaraman Lab filter
- Lippincott-Schwartz Lab (1) Apply Lippincott-Schwartz Lab filter
- Looger Lab (2) Apply Looger Lab filter
- O'Shea Lab (1) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Reiser Lab (15) Apply Reiser Lab filter
- Riddiford Lab (1) Apply Riddiford Lab filter
- Romani Lab (1) Apply Romani Lab filter
- Rubin Lab (143) Apply Rubin Lab filter
- Saalfeld Lab (4) Apply Saalfeld Lab filter
- Scheffer Lab (7) Apply Scheffer Lab filter
- Schreiter Lab (1) Apply Schreiter Lab filter
- Simpson Lab (3) Apply Simpson Lab filter
- Singer Lab (1) Apply Singer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Stern Lab (1) Apply Stern Lab filter
- Svoboda Lab (3) Apply Svoboda Lab filter
- Tjian Lab (1) Apply Tjian Lab filter
- Truman Lab (4) Apply Truman Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
- Turner Lab (5) Apply Turner Lab filter
- Zuker Lab (1) Apply Zuker Lab filter
Associated Project Team
- CellMap (1) Apply CellMap filter
- Fly Functional Connectome (4) Apply Fly Functional Connectome filter
- Fly Olympiad (3) Apply Fly Olympiad filter
- FlyEM (11) Apply FlyEM filter
- FlyLight (20) Apply FlyLight filter
- GENIE (1) Apply GENIE filter
- Transcription Imaging (1) Apply Transcription Imaging filter
Publication Date
- 2025 (4) Apply 2025 filter
- 2024 (4) Apply 2024 filter
- 2023 (5) Apply 2023 filter
- 2022 (1) Apply 2022 filter
- 2021 (4) Apply 2021 filter
- 2020 (9) Apply 2020 filter
- 2019 (6) Apply 2019 filter
- 2018 (7) Apply 2018 filter
- 2017 (15) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (16) Apply 2015 filter
- 2014 (9) Apply 2014 filter
- 2013 (5) Apply 2013 filter
- 2012 (8) Apply 2012 filter
- 2011 (4) Apply 2011 filter
- 2010 (4) Apply 2010 filter
- 2009 (2) Apply 2009 filter
- 2008 (4) Apply 2008 filter
- 2007 (2) Apply 2007 filter
- 2006 (1) Apply 2006 filter
- 2002 (1) Apply 2002 filter
- 2000 (2) Apply 2000 filter
- 1999 (1) Apply 1999 filter
- 1997 (1) Apply 1997 filter
- 1995 (2) Apply 1995 filter
- 1994 (2) Apply 1994 filter
- 1993 (2) Apply 1993 filter
- 1992 (1) Apply 1992 filter
- 1991 (2) Apply 1991 filter
- 1990 (3) Apply 1990 filter
- 1989 (2) Apply 1989 filter
- 1987 (2) Apply 1987 filter
- 1986 (1) Apply 1986 filter
- 1985 (1) Apply 1985 filter
- 1984 (1) Apply 1984 filter
- 1983 (1) Apply 1983 filter
- 1982 (2) Apply 1982 filter
- 1981 (1) Apply 1981 filter
- 1979 (1) Apply 1979 filter
- 1973 (1) Apply 1973 filter
Type of Publication
143 Publications
Showing 121-130 of 143 resultsNull mutations of glass specifically remove photoreceptor cells, leaving other cell types intact. We have isolated the glass gene and have shown that its transcript encodes a putative protein of 604 amino acids with five zinc-fingers. The glass product may be a transcription factor required for the development of a single neuronal cell type.
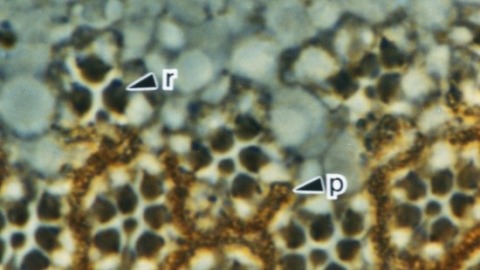
Glia play crucial roles in the development and homeostasis of the nervous system. While the GLIA in the Drosophila embryo have been well characterized, their study in the adult nervous system has been limited. Here, we present a detailed description of the glia in the adult nervous system, based on the analysis of some 500 glial drivers we identified within a collection of synthetic GAL4 lines. We find that glia make up ∼10% of the cells in the nervous system and envelop all compartments of neurons (soma, dendrites, axons) as well as the nervous system as a whole. Our morphological analysis suggests a set of simple rules governing the morphogenesis of glia and their interactions with other cells. All glial subtypes minimize contact with their glial neighbors but maximize their contact with neurons and adapt their macromorphology and micromorphology to the neuronal entities they envelop. Finally, glial cells show no obvious spatial organization or registration with neuronal entities. Our detailed description of all glial subtypes and their regional specializations, together with the powerful genetic toolkit we provide, will facilitate the functional analysis of glia in the mature nervous system. GLIA 2017.
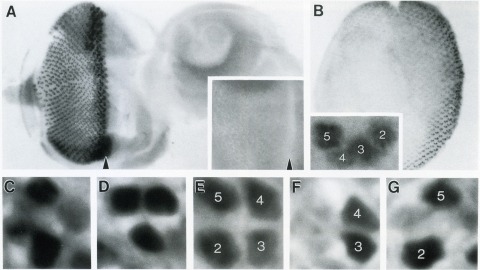
The Drosophila homeo box gene rough is required in photoreceptor cells R2 and R5 for normal eye development. We show here that rough protein expression is limited to a subset of cells in the developing retina where it is transiently expressed for 30-60 hr. The rough protein is first expressed broadly in the morphogenetic furrow but is rapidly restricted to the R2, R3, R4, and R5 precursor cells. Ubiquitous expression of rough under the control of the hsp70 promoter in third-instar larvae suppresses the initial steps of ommatidial assembly. Structures derived from other imaginal discs are not affected. Ectopic expression of rough in the R7 precursor, through the use of the sevenless promoter, causes this cell to develop into an R1-6 photoreceptor subtype; however, this cell still requires sevenless function for its neural differentiation. Taken together with previous analyses of the rough mutant phenotype, these results suggest that the normal role of rough is to establish the unique cell identity of photoreceptors R2 and R5.
Large scientific projects in genomics and astronomy are influential not because they answer any single question but because they enable investigation of continuously arising new questions from the same data-rich sources. Advances in automated mapping of the brain's synaptic connections (connectomics) suggest that the complicated circuits underlying brain function are ripe for analysis. We discuss benefits of mapping a mouse brain at the level of synapses.
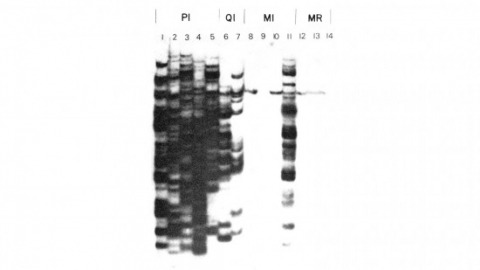
We have shown previously that four of five white mutant alleles arising in P-M dysgenic hybrids result from the insertion of strongly homologous DNA sequence elements. We have named these P elements. We report that P elements are present in 30-50 copies per haploid genome in all P strains examined and apparently are missing entirely from all M strains examined, with one exception. Furthermore, members of the P family apparently transpose frequently in P-M dysgenic hybrids; chromosomes descendant from P-M dysgenic hybrids frequently show newly acquired P elements. Finally, the strain-specific breakpoint hotspots for the rearrangement of the pi 2 P X chromosome occurring in P-M dysgenic hybrids are apparently sites of residence of P elements. These observations strongly support the P factor hypothesis for the mechanistic basis of P-M hybrid dysgenesis.
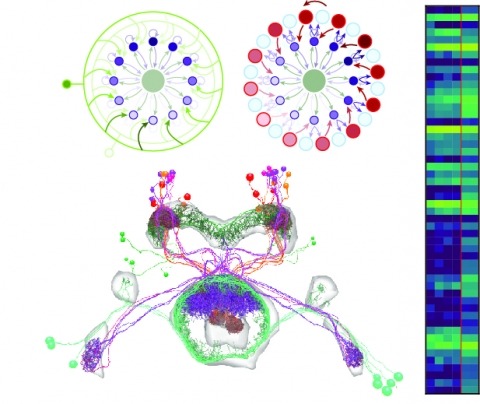
Neural representations of head direction (HD) have been discovered in many species. Theoretical work has proposed that the dynamics associated with these representations are generated, maintained, and updated by recurrent network structures called ring attractors. We evaluated this theorized structure-function relationship by performing electron-microscopy-based circuit reconstruction and RNA profiling of identified cell types in the HD system of Drosophila melanogaster. We identified motifs that have been hypothesized to maintain the HD representation in darkness, update it when the animal turns, and tether it to visual cues. Functional studies provided support for the proposed roles of individual excitatory or inhibitory circuit elements in shaping activity. We also discovered recurrent connections between neuronal arbors with mixed pre- and postsynaptic specializations. Our results confirm that the Drosophila HD network contains the core components of a ring attractor while also revealing unpredicted structural features that might enhance the network's computational power.
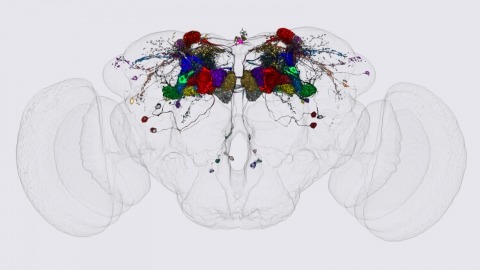
We identified the neurons comprising the Drosophila mushroom body (MB), an associative center in invertebrate brains, and provide a comprehensive map describing their potential connections. Each of the 21 MB output neuron (MBON) types elaborates segregated dendritic arbors along the parallel axons of ∼2000 Kenyon cells, forming 15 compartments that collectively tile the MB lobes. MBON axons project to five discrete neuropils outside of the MB and three MBON types form a feedforward network in the lobes. Each of the 20 dopaminergic neuron (DAN) types projects axons to one, or at most two, of the MBON compartments. Convergence of DAN axons on compartmentalized Kenyon cell-MBON synapses creates a highly ordered unit that can support learning to impose valence on sensory representations. The elucidation of the complement of neurons of the MB provides a comprehensive anatomical substrate from which one can infer a functional logic of associative olfactory learning and memory.
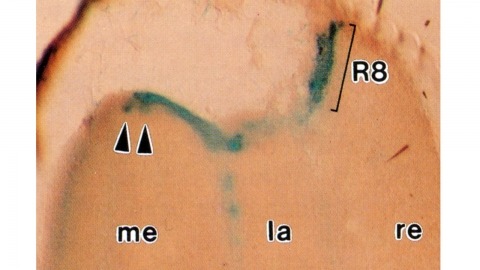
Histological staining of wild-type and sevenless transgenic Drosophila melanogaster bearing Rh3-lacZ fusion genes permits the selective visualization of polarization-sensitive R7 and R8 photoreceptor cells located along the dorsal anterior eye margin. Diffusion of beta-galactosidase throughout these cells reveals that they project long axons to the two most peripheral synaptic target rows of the dorsal posterior medulla, defining a specialized marginal zone of this optic lobe. Comparison of the staining patterns of marginal and nonmarginal Rh3-lacZ-expressing photoreceptor cells in the same histological preparations suggest that the marginal cells possess morphologically specialized axons and synaptic terminals. These findings are discussed with reference to the neuroanatomy of the corresponding dorsal marginal eye and optic lobe regions of the larger dipterans Musca and Calliphora, and in relation to the ability of Drosophila to orient to polarized light.
Drosophila melanogaster plays an important role in molecular, genetic, and genomic studies of heredity, development, metabolism, behavior, and human disease. The initial reference genome sequence reported more than a decade ago had a profound impact on progress in Drosophila research, and improving the accuracy and completeness of this sequence continues to be important to further progress. We previously described improvement of the 117-Mb sequence in the euchromatic portion of the genome and 21 Mb in the heterochromatic portion, using a whole-genome shotgun assembly, BAC physical mapping, and clone-based finishing. Here, we report an improved reference sequence of the single-copy and middle-repetitive regions of the genome, produced using cytogenetic mapping to mitotic and polytene chromosomes, clone-based finishing and BAC fingerprint verification, ordering of scaffolds by alignment to cDNA sequences, incorporation of other map and sequence data, and validation by whole-genome optical restriction mapping. These data substantially improve the accuracy and completeness of the reference sequence and the order and orientation of sequence scaffolds into chromosome arm assemblies. Representation of the Y chromosome and other heterochromatic regions is particularly improved. The new 143.9-Mb reference sequence, designated Release 6, effectively exhausts clone-based technologies for mapping and sequencing. Highly repeat-rich regions, including large satellite blocks and functional elements such as the ribosomal RNA genes and the centromeres, are largely inaccessible to current sequencing and assembly methods and remain poorly represented. Further significant improvements will require sequencing technologies that do not depend on molecular cloning and that produce very long reads.