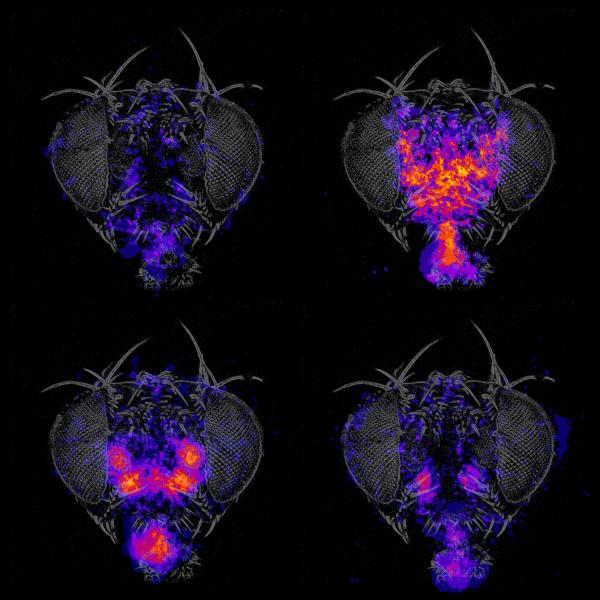
We use genetic tools and screening strategies to identify the specific neurons necessary and sufficient to control grooming and feeding, behaviors which were chosen for their sequential progression and cue integration properties. Detailed mapping of the circuits that govern these specific behaviors in flies may reveal common architectural principles that help explain how human brains produce a diverse repertoire of appropriate behavior in response to internal state and external stimuli.
Some behaviors seem like simple reflexes – sugar on a fly’s leg triggers extension of the mouthparts, and dust on a fly’s back evokes leg sweeps. Sensory inputs and motor outputs are critical.
But a fly only extends its mouthparts when it is hungry, and when the sweet taste is not accompanied by a bitter one. The neurons that integrate several sensory inputs and internal state information have not yet been found. Activating the mechanosensory neurons at the base of bristles can trigger a scratch reflex, but activating many bristles results in an ordered series of leg sweeps and rubs designed to remove debris from the eyes and head, followed by the wings and body. The motor neurons required to perform grooming movements may also be critical for walking. How they are organized to perform different tasks in different contexts is not known.
The fly brain has a lot of terra incognita – areas whose behavioral functions are undiscovered. The anatomical regions relevant for many behaviors are not known. The way a given behavior can be disrupted is often informative, even if the neural circuit or cause is not yet known. We identify the neurons needed for particular behaviors, rather than the genes, using the plethora of genetic tools available in Drosophila to target, manipulate, and image neurons. We also mirror the tradition of unbiased genetics screens in that we test the potential role of many different groups of neurons (using the large Rubin GAL4 collection and others) and allow the results to suggest which brain regions are relevant. This approach provides entry points into the circuits, from which we can then search anatomically for potentially connected neurons.
Grooming
Flies remove dust from their bodies in an orderly way, starting from their eyes and proceeding to their heads, wings, and abdomens. This progression is likely influenced by mechanosensory feedback. The data from our observation of grooming behavior in normal flies and in flies where the activity of different groups of neurons has been altered supports the idea that grooming is made up of small reflex modules or subroutines linked together. We are identifying the neurons that control the reflex components and the neurons that coordinate them to produce the grooming behavioral sequence. Contrasting the way behavioral sequences are produced for courtship and grooming may be particularly informative.
Feeding and Proboscis Extension
Flies eat when they are hungry and good food is available. Generally they do not eat more than they need. We want to identify neurons that can stimulate feeding in sated flies to map the brain areas that detect metabolic state and encode hunger. We will explore where the external sensory cues (sweet and bitter taste) are integrated with the internal metabolic state cues to activate the motor program of the proboscis extension reflex (PER).
PER is activated during feeding and drinking, but it can also be evoked during courtship or in response to aversive stimuli. We are investigating how these sensory stimuli converge on what we think is a common program to control this motor behavior. The motor program itself contains 32 motor neurons and 12 muscles that function in a coordinated manner to extend and retract the proboscis. While the motor neurons are known by traditional anatomical techniques, the ability to alter activity in particular neurons is recent and allows us to study how this motor program is generated.
Courtship Song Circuits
Male Drosophila melanogaster produce courtship song by vibrating their wings at 160Hz (sine) and 260Hz (pulse). The male-specific isoform of Fruitless marks many of the neurons necessary and sufficient for this behavior, but other components of the song circuit remain elusive. Although the basic nature of the song is innate, some aspects may be modified by experience, sensory feedback, or social context. We are analyzing the song structure and identifying neurons that drive song production as another example of how neural circuits produce behavioral sequences.
Tool Development: dBrainbow and BrainAligner
We are screening for neurons necessary and sufficient for behaviors, not the genes. This requires tools to manipulate specific populations of neurons. Initially we used enhancer trapping to generate collections of GAL4, GAL80, and LexA lines that we use to target neural activators and inhibitors to different brain regions. Now we primarily use the defined enhancer GAL4 and split-GAL4 collections established by the Rubin lab. We are building additional reagents to manipulate different types of neurons and are developing a web site to display and query the expression patterns of our lines.
We adapted the vertebrate Brainbow technique for labeling neurons in different colors to visualize lineages and individual neurons within our GAL4 expression patterns (Nature Methods 2011).
In collaboration with Hanchuan Peng and Gene Myers, we developed the BrainAligner for automatically registering confocal images of fly brains onto a common coordinate system. This allows us to generate an atlas of all our GAL4 expression patterns to identify lines that target particular brain regions. We use this to make predictions about anatomical correlation with behavioral function and connectivity (Submitted 2010).
Collaborations
The Simpson lab participates in the Drosophila Interests Group (a joint lab meeting of the 10 fly labs at Janelia) and the Behavior Interest Group (an occasional meeting of the Janelia neuroethology community). We are also part of two project teams, FlyLight and the FlyOlympiad. There is a coordinated effort to “Understand the Fly Brain” at Janelia, using light microscopy, electron microscopy, and large-scale quantitative behavioral analysis. My lab and I participate actively in this joint effort.
We understand that functional and anatomical mapping of the fly brain is an ambitious task, and we hope that our efforts will contribute to the larger community-wide endeavor. The anatomical images, circuit maps, and genetic tools we make can be used to address a broad range of scientific questions well beyond those we focus on in one lab. We feel strongly that tools are improved by sharing and we freely distribute the reagents we make. We look forward to hearing from anyone interested in our research.
Collaborators
- Gerald Rubin
- Loren Looger
- Gene Myers
- Hanchuan Peng
Contact
Contact me at: jhsimpson@lifesci.ucsb.edu